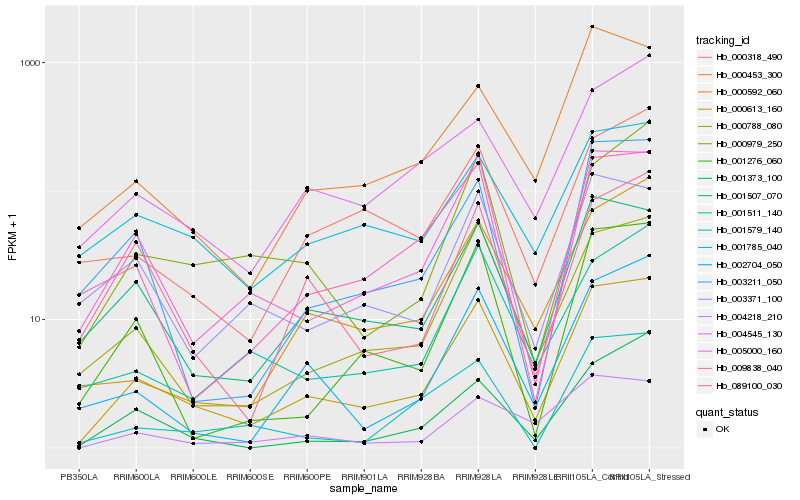
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000613_160 |
0.0 |
- |
- |
coated vesicle membrane protein, putative [Ricinus communis] |
| 2 |
Hb_000788_080 |
0.1206280657 |
- |
- |
PREDICTED: uncharacterized protein LOC105629703 isoform X1 [Jatropha curcas] |
| 3 |
Hb_005000_160 |
0.12326921 |
- |
- |
PREDICTED: uncharacterized protein LOC105637882 [Jatropha curcas] |
| 4 |
Hb_001507_070 |
0.135367675 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_002704_050 |
0.142954923 |
- |
- |
cysteine-type peptidase, putative [Ricinus communis] |
| 6 |
Hb_001276_060 |
0.1430549152 |
- |
- |
hypothetical protein POPTR_0015s00420g [Populus trichocarpa] |
| 7 |
Hb_000979_250 |
0.1517201876 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_009838_040 |
0.1538769609 |
- |
- |
hypothetical protein JCGZ_12866 [Jatropha curcas] |
| 9 |
Hb_001511_140 |
0.1543814111 |
- |
- |
PREDICTED: RING-H2 finger protein ATL66 [Jatropha curcas] |
| 10 |
Hb_003211_050 |
0.154841267 |
- |
- |
PREDICTED: RNA-binding protein 24-B isoform X1 [Jatropha curcas] |
| 11 |
Hb_000592_060 |
0.1551780448 |
- |
- |
PREDICTED: ras-related protein RABA6a [Populus euphratica] |
| 12 |
Hb_001373_100 |
0.1567361606 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_004218_210 |
0.1573349813 |
transcription factor |
TF Family: Orphans |
hypothetical protein JCGZ_24525 [Jatropha curcas] |
| 14 |
Hb_000453_300 |
0.1587178179 |
- |
- |
latex cystatin [Hevea brasiliensis] |
| 15 |
Hb_001579_140 |
0.1590789174 |
- |
- |
PREDICTED: probable inactive poly [ADP-ribose] polymerase SRO2 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000318_490 |
0.1602182885 |
- |
- |
PREDICTED: mitochondrial import receptor subunit TOM6 homolog [Jatropha curcas] |
| 17 |
Hb_001785_040 |
0.1611126674 |
- |
- |
cytochrome b5 isoform Cb5-A [Vernicia fordii] |
| 18 |
Hb_089100_030 |
0.1624235721 |
- |
- |
- |
| 19 |
Hb_004545_130 |
0.1626198998 |
- |
- |
uncharacterized protein LOC100305465 [Glycine max] |
| 20 |
Hb_003371_100 |
0.1627331458 |
transcription factor |
TF Family: HB |
homeobox protein, putative [Ricinus communis] |