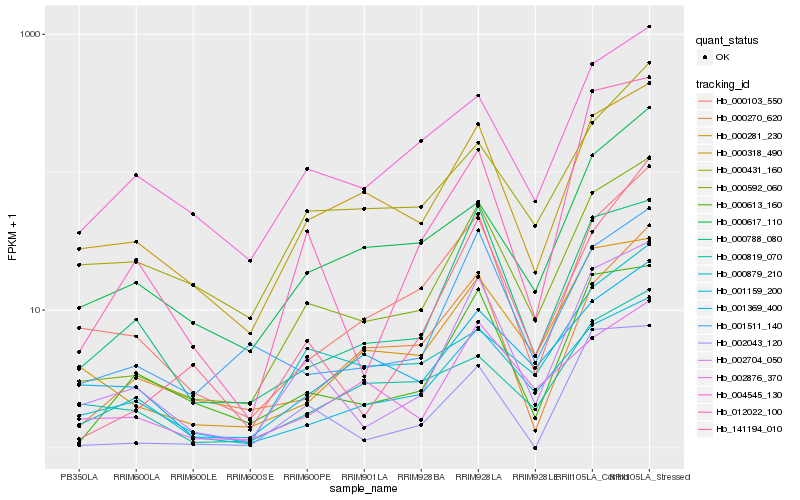
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000592_060 |
0.0 |
- |
- |
PREDICTED: ras-related protein RABA6a [Populus euphratica] |
| 2 |
Hb_002704_050 |
0.1119059543 |
- |
- |
cysteine-type peptidase, putative [Ricinus communis] |
| 3 |
Hb_000431_160 |
0.1275324865 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit gamma-like [Jatropha curcas] |
| 4 |
Hb_000318_490 |
0.1287792472 |
- |
- |
PREDICTED: mitochondrial import receptor subunit TOM6 homolog [Jatropha curcas] |
| 5 |
Hb_000103_550 |
0.1373234565 |
- |
- |
PREDICTED: guanine nucleotide-binding protein subunit gamma 2 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000281_230 |
0.1403890752 |
- |
- |
PREDICTED: probable lysine--tRNA ligase, cytoplasmic isoform X2 [Jatropha curcas] |
| 7 |
Hb_004545_130 |
0.1411090364 |
- |
- |
uncharacterized protein LOC100305465 [Glycine max] |
| 8 |
Hb_001159_200 |
0.1454027235 |
- |
- |
PREDICTED: clp protease-related protein At4g12060, chloroplastic-like [Jatropha curcas] |
| 9 |
Hb_001511_140 |
0.1478118628 |
- |
- |
PREDICTED: RING-H2 finger protein ATL66 [Jatropha curcas] |
| 10 |
Hb_000617_110 |
0.1533316487 |
- |
- |
translationally controlled tumor protein [Hevea brasiliensis] |
| 11 |
Hb_000613_160 |
0.1551780448 |
- |
- |
coated vesicle membrane protein, putative [Ricinus communis] |
| 12 |
Hb_000879_210 |
0.1552284311 |
- |
- |
hypothetical protein POPTR_0014s01620g [Populus trichocarpa] |
| 13 |
Hb_141194_010 |
0.1570946995 |
- |
- |
PREDICTED: protein ZINC INDUCED FACILITATOR 1-like isoform X2 [Populus euphratica] |
| 14 |
Hb_000788_080 |
0.1580695003 |
- |
- |
PREDICTED: uncharacterized protein LOC105629703 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000270_620 |
0.1618209928 |
- |
- |
PREDICTED: succinate dehydrogenase subunit 7B, mitochondrial-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_002876_370 |
0.1632420798 |
- |
- |
hypothetical protein B456_013G140400 [Gossypium raimondii] |
| 17 |
Hb_001369_400 |
0.1659631131 |
- |
- |
PREDICTED: uncharacterized protein LOC105648609 [Jatropha curcas] |
| 18 |
Hb_012022_100 |
0.1665912532 |
- |
- |
CPRD49 family protein [Populus trichocarpa] |
| 19 |
Hb_002043_120 |
0.1720425691 |
- |
- |
protein with unknown function [Ricinus communis] |
| 20 |
Hb_000819_070 |
0.1733482904 |
- |
- |
PREDICTED: dolichol-phosphate mannosyltransferase subunit 1 [Jatropha curcas] |