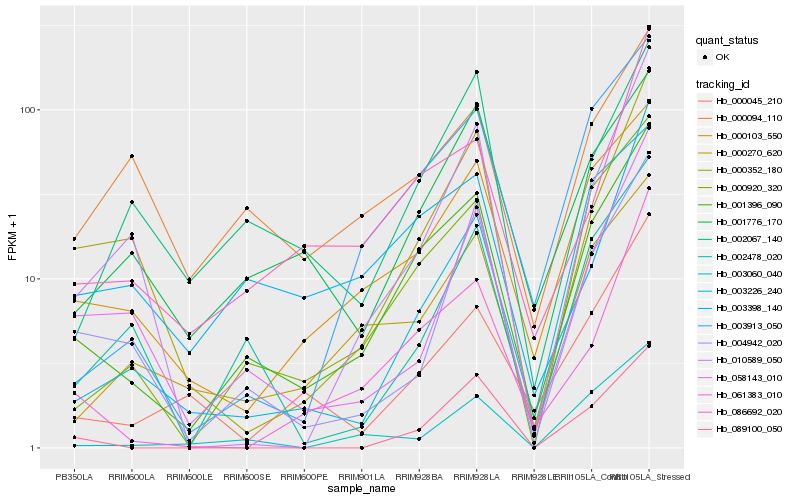
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001396_090 |
0.0 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 75 [Jatropha curcas] |
| 2 |
Hb_000920_320 |
0.1196130031 |
transcription factor |
TF Family: Orphans |
hypothetical protein JCGZ_24525 [Jatropha curcas] |
| 3 |
Hb_000103_550 |
0.1337336274 |
- |
- |
PREDICTED: guanine nucleotide-binding protein subunit gamma 2 isoform X1 [Jatropha curcas] |
| 4 |
Hb_003913_050 |
0.1422456949 |
- |
- |
60S ribosomal protein L27B [Hevea brasiliensis] |
| 5 |
Hb_000270_620 |
0.1568665418 |
- |
- |
PREDICTED: succinate dehydrogenase subunit 7B, mitochondrial-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_061383_010 |
0.1640512477 |
- |
- |
beta-1,3-glucanase [Hevea brasiliensis] |
| 7 |
Hb_001776_170 |
0.1647262501 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_086692_020 |
0.1735120911 |
- |
- |
PREDICTED: protein kinase APK1B, chloroplastic-like [Jatropha curcas] |
| 9 |
Hb_002478_020 |
0.1746635095 |
- |
- |
hypothetical protein POPTR_0010s19120g [Populus trichocarpa] |
| 10 |
Hb_004942_020 |
0.1800873282 |
- |
- |
Potassium transporter, putative [Ricinus communis] |
| 11 |
Hb_000352_180 |
0.1813808339 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_058143_010 |
0.1821648802 |
- |
- |
hypothetical protein JCGZ_01992 [Jatropha curcas] |
| 13 |
Hb_000045_210 |
0.1834019126 |
- |
- |
- |
| 14 |
Hb_003060_040 |
0.1856127717 |
transcription factor |
TF Family: MIKC |
PREDICTED: agamous-like MADS-box protein AGL15 [Jatropha curcas] |
| 15 |
Hb_003226_240 |
0.1866669031 |
- |
- |
PREDICTED: caffeoylshikimate esterase [Jatropha curcas] |
| 16 |
Hb_010589_050 |
0.1909882248 |
- |
- |
- |
| 17 |
Hb_003398_140 |
0.1929618503 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 18 |
Hb_002067_140 |
0.1932288991 |
- |
- |
- |
| 19 |
Hb_089100_050 |
0.193484626 |
- |
- |
- |
| 20 |
Hb_000094_110 |
0.198118241 |
- |
- |
conserved hypothetical protein [Ricinus communis] |