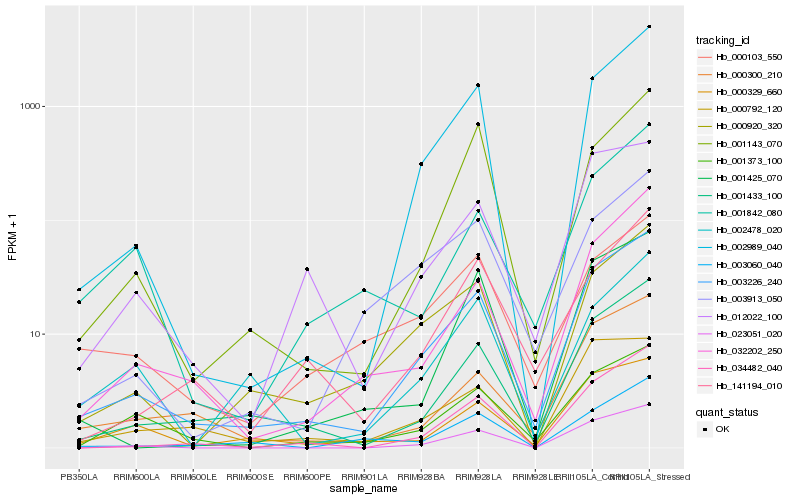
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003226_240 |
0.0 |
- |
- |
PREDICTED: caffeoylshikimate esterase [Jatropha curcas] |
| 2 |
Hb_002989_040 |
0.1005207247 |
- |
- |
hypothetical protein 1 [Hevea brasiliensis] |
| 3 |
Hb_001433_100 |
0.1062591419 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 3 isoform X1 [Jatropha curcas] |
| 4 |
Hb_034482_040 |
0.1124343698 |
- |
- |
aspartate kinase, partial [Francisella tularensis] |
| 5 |
Hb_023051_020 |
0.1159507025 |
- |
- |
- |
| 6 |
Hb_000920_320 |
0.1248101063 |
transcription factor |
TF Family: Orphans |
hypothetical protein JCGZ_24525 [Jatropha curcas] |
| 7 |
Hb_032202_250 |
0.1315722629 |
- |
- |
PREDICTED: F-box protein PP2-A13 [Jatropha curcas] |
| 8 |
Hb_000300_210 |
0.1363481802 |
- |
- |
gd2b, putative [Ricinus communis] |
| 9 |
Hb_012022_100 |
0.1444020362 |
- |
- |
CPRD49 family protein [Populus trichocarpa] |
| 10 |
Hb_001425_070 |
0.1490027691 |
- |
- |
- |
| 11 |
Hb_001143_070 |
0.1496923468 |
- |
- |
Acyl-CoA N-acyltransferases (NAT) superfamily protein isoform 1 [Theobroma cacao] |
| 12 |
Hb_001373_100 |
0.1502248067 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_003913_050 |
0.1526108781 |
- |
- |
60S ribosomal protein L27B [Hevea brasiliensis] |
| 14 |
Hb_141194_010 |
0.1531146816 |
- |
- |
PREDICTED: protein ZINC INDUCED FACILITATOR 1-like isoform X2 [Populus euphratica] |
| 15 |
Hb_000329_660 |
0.15700735 |
- |
- |
PREDICTED: carotenoid cleavage dioxygenase 7, chloroplastic [Jatropha curcas] |
| 16 |
Hb_003060_040 |
0.161369143 |
transcription factor |
TF Family: MIKC |
PREDICTED: agamous-like MADS-box protein AGL15 [Jatropha curcas] |
| 17 |
Hb_001842_080 |
0.1680514094 |
- |
- |
- |
| 18 |
Hb_000792_120 |
0.1702264422 |
- |
- |
PREDICTED: mitochondrial uncoupling protein 1 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000103_550 |
0.1715377003 |
- |
- |
PREDICTED: guanine nucleotide-binding protein subunit gamma 2 isoform X1 [Jatropha curcas] |
| 20 |
Hb_002478_020 |
0.1718998439 |
- |
- |
hypothetical protein POPTR_0010s19120g [Populus trichocarpa] |