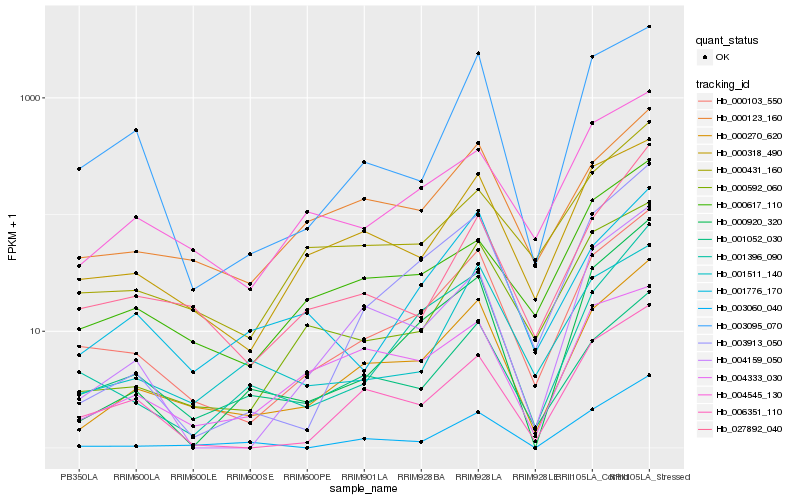
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000270_620 |
0.0 |
- |
- |
PREDICTED: succinate dehydrogenase subunit 7B, mitochondrial-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_000103_550 |
0.1070424381 |
- |
- |
PREDICTED: guanine nucleotide-binding protein subunit gamma 2 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000123_160 |
0.1280040845 |
- |
- |
PREDICTED: 40S ribosomal protein S15a-1 [Jatropha curcas] |
| 4 |
Hb_000318_490 |
0.137833253 |
- |
- |
PREDICTED: mitochondrial import receptor subunit TOM6 homolog [Jatropha curcas] |
| 5 |
Hb_004159_050 |
0.1453041156 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000920_320 |
0.1454999756 |
transcription factor |
TF Family: Orphans |
hypothetical protein JCGZ_24525 [Jatropha curcas] |
| 7 |
Hb_004545_130 |
0.1519361816 |
- |
- |
uncharacterized protein LOC100305465 [Glycine max] |
| 8 |
Hb_000431_160 |
0.1542378723 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit gamma-like [Jatropha curcas] |
| 9 |
Hb_001396_090 |
0.1568665418 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 75 [Jatropha curcas] |
| 10 |
Hb_003913_050 |
0.1570596059 |
- |
- |
60S ribosomal protein L27B [Hevea brasiliensis] |
| 11 |
Hb_000617_110 |
0.1573142219 |
- |
- |
translationally controlled tumor protein [Hevea brasiliensis] |
| 12 |
Hb_003060_040 |
0.1576340966 |
transcription factor |
TF Family: MIKC |
PREDICTED: agamous-like MADS-box protein AGL15 [Jatropha curcas] |
| 13 |
Hb_027892_040 |
0.1580440294 |
- |
- |
PREDICTED: proteasome subunit alpha type-6 [Jatropha curcas] |
| 14 |
Hb_006351_110 |
0.1582613969 |
- |
- |
- |
| 15 |
Hb_004333_030 |
0.158880727 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 26A [Jatropha curcas] |
| 16 |
Hb_001776_170 |
0.159005327 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001052_030 |
0.1590200691 |
- |
- |
PREDICTED: protein TIC 20-IV, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000592_060 |
0.1618209928 |
- |
- |
PREDICTED: ras-related protein RABA6a [Populus euphratica] |
| 19 |
Hb_001511_140 |
0.1619258331 |
- |
- |
PREDICTED: RING-H2 finger protein ATL66 [Jatropha curcas] |
| 20 |
Hb_003095_070 |
0.1644165816 |
- |
- |
hypothetical protein POPTR_0009s11230g [Populus trichocarpa] |