| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
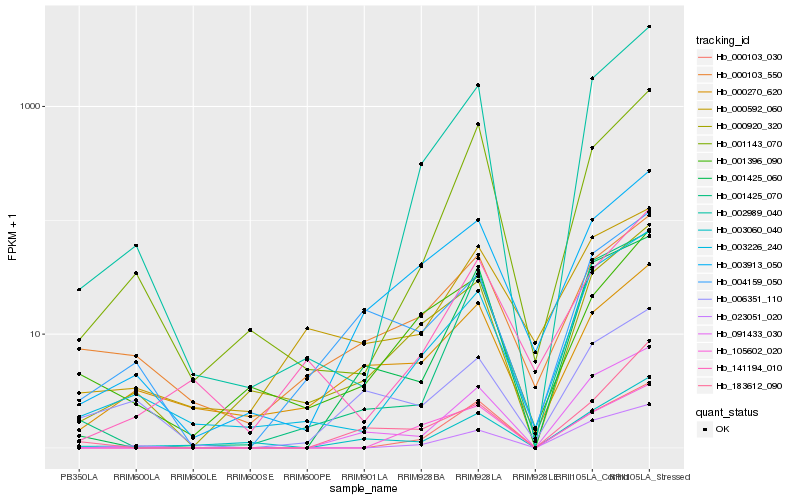
Hb_003913_050 |
0.0 |
- |
- |
60S ribosomal protein L27B [Hevea brasiliensis] |
| 2 |
Hb_000920_320 |
0.1180997228 |
transcription factor |
TF Family: Orphans |
hypothetical protein JCGZ_24525 [Jatropha curcas] |
| 3 |
Hb_001396_090 |
0.1422456949 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 75 [Jatropha curcas] |
| 4 |
Hb_000103_550 |
0.1460439774 |
- |
- |
PREDICTED: guanine nucleotide-binding protein subunit gamma 2 isoform X1 [Jatropha curcas] |
| 5 |
Hb_003226_240 |
0.1526108781 |
- |
- |
PREDICTED: caffeoylshikimate esterase [Jatropha curcas] |
| 6 |
Hb_004159_050 |
0.1569995267 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000270_620 |
0.1570596059 |
- |
- |
PREDICTED: succinate dehydrogenase subunit 7B, mitochondrial-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_002989_040 |
0.1600750519 |
- |
- |
hypothetical protein 1 [Hevea brasiliensis] |
| 9 |
Hb_091433_030 |
0.1648729058 |
- |
- |
- |
| 10 |
Hb_003060_040 |
0.1668592993 |
transcription factor |
TF Family: MIKC |
PREDICTED: agamous-like MADS-box protein AGL15 [Jatropha curcas] |
| 11 |
Hb_105602_020 |
0.1729416724 |
- |
- |
PREDICTED: 60S ribosomal protein L19-1-like [Populus euphratica] |
| 12 |
Hb_000592_060 |
0.174940505 |
- |
- |
PREDICTED: ras-related protein RABA6a [Populus euphratica] |
| 13 |
Hb_183612_090 |
0.1793621061 |
- |
- |
- |
| 14 |
Hb_001425_060 |
0.1796034806 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_141194_010 |
0.1855834473 |
- |
- |
PREDICTED: protein ZINC INDUCED FACILITATOR 1-like isoform X2 [Populus euphratica] |
| 16 |
Hb_006351_110 |
0.1875876503 |
- |
- |
- |
| 17 |
Hb_001143_070 |
0.1890996414 |
- |
- |
Acyl-CoA N-acyltransferases (NAT) superfamily protein isoform 1 [Theobroma cacao] |
| 18 |
Hb_000103_030 |
0.1962606244 |
- |
- |
hypothetical protein VITISV_021700 [Vitis vinifera] |
| 19 |
Hb_023051_020 |
0.1990493632 |
- |
- |
- |
| 20 |
Hb_001425_070 |
0.1992749793 |
- |
- |
- |