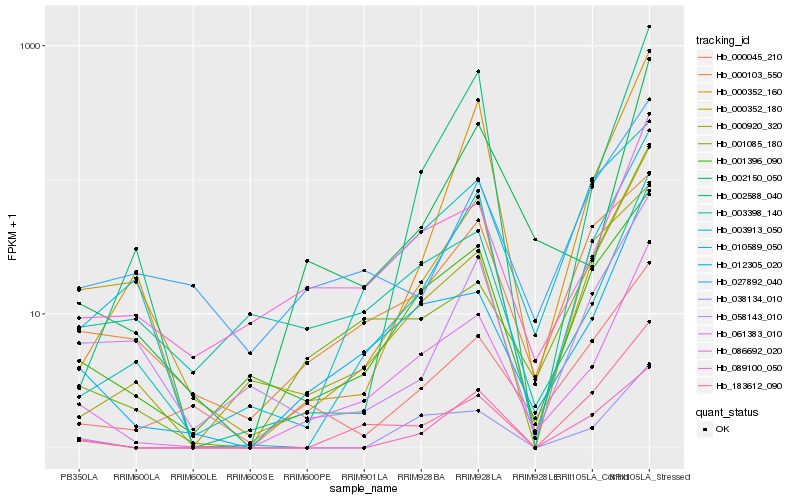
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_061383_010 |
0.0 |
- |
- |
beta-1,3-glucanase [Hevea brasiliensis] |
| 2 |
Hb_012305_020 |
0.0914366961 |
- |
- |
beta-1,3-glucanase [Hevea brasiliensis] |
| 3 |
Hb_086692_020 |
0.144493189 |
- |
- |
PREDICTED: protein kinase APK1B, chloroplastic-like [Jatropha curcas] |
| 4 |
Hb_002150_050 |
0.1606890214 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001396_090 |
0.1640512477 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 75 [Jatropha curcas] |
| 6 |
Hb_038134_010 |
0.1746553087 |
- |
- |
- |
| 7 |
Hb_183612_090 |
0.1767573761 |
- |
- |
- |
| 8 |
Hb_001085_180 |
0.1833506644 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_010589_050 |
0.1931969837 |
- |
- |
- |
| 10 |
Hb_000352_180 |
0.1988919213 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000045_210 |
0.2001401669 |
- |
- |
- |
| 12 |
Hb_003913_050 |
0.2092353901 |
- |
- |
60S ribosomal protein L27B [Hevea brasiliensis] |
| 13 |
Hb_002588_040 |
0.2114041121 |
- |
- |
Pathogenesis-related protein 1 [Theobroma cacao] |
| 14 |
Hb_000352_160 |
0.2209514176 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000103_550 |
0.2221350488 |
- |
- |
PREDICTED: guanine nucleotide-binding protein subunit gamma 2 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000920_320 |
0.2243155352 |
transcription factor |
TF Family: Orphans |
hypothetical protein JCGZ_24525 [Jatropha curcas] |
| 17 |
Hb_089100_050 |
0.2252891156 |
- |
- |
- |
| 18 |
Hb_058143_010 |
0.2255620625 |
- |
- |
hypothetical protein JCGZ_01992 [Jatropha curcas] |
| 19 |
Hb_003398_140 |
0.2259749109 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 20 |
Hb_027892_040 |
0.226475163 |
- |
- |
PREDICTED: proteasome subunit alpha type-6 [Jatropha curcas] |