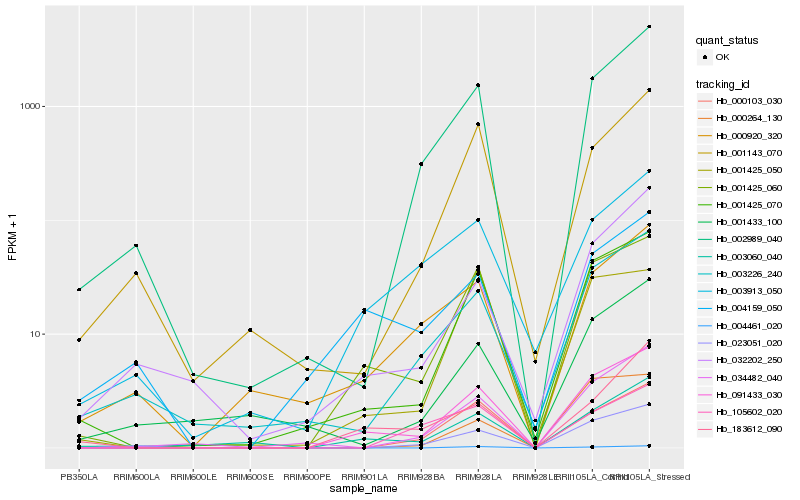
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_091433_030 |
0.0 |
- |
- |
- |
| 2 |
Hb_001425_060 |
0.0700268792 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001425_070 |
0.114152497 |
- |
- |
- |
| 4 |
Hb_003060_040 |
0.154694388 |
transcription factor |
TF Family: MIKC |
PREDICTED: agamous-like MADS-box protein AGL15 [Jatropha curcas] |
| 5 |
Hb_002989_040 |
0.1548355906 |
- |
- |
hypothetical protein 1 [Hevea brasiliensis] |
| 6 |
Hb_023051_020 |
0.1557837308 |
- |
- |
- |
| 7 |
Hb_000103_030 |
0.1625334746 |
- |
- |
hypothetical protein VITISV_021700 [Vitis vinifera] |
| 8 |
Hb_003913_050 |
0.1648729058 |
- |
- |
60S ribosomal protein L27B [Hevea brasiliensis] |
| 9 |
Hb_034482_040 |
0.1652600639 |
- |
- |
aspartate kinase, partial [Francisella tularensis] |
| 10 |
Hb_003226_240 |
0.1720509331 |
- |
- |
PREDICTED: caffeoylshikimate esterase [Jatropha curcas] |
| 11 |
Hb_004159_050 |
0.1793611403 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000920_320 |
0.1796562832 |
transcription factor |
TF Family: Orphans |
hypothetical protein JCGZ_24525 [Jatropha curcas] |
| 13 |
Hb_183612_090 |
0.181020282 |
- |
- |
- |
| 14 |
Hb_001143_070 |
0.1875411803 |
- |
- |
Acyl-CoA N-acyltransferases (NAT) superfamily protein isoform 1 [Theobroma cacao] |
| 15 |
Hb_032202_250 |
0.1919572915 |
- |
- |
PREDICTED: F-box protein PP2-A13 [Jatropha curcas] |
| 16 |
Hb_004461_020 |
0.1928714557 |
- |
- |
PREDICTED: uncharacterized protein LOC104105697 [Nicotiana tomentosiformis] |
| 17 |
Hb_001425_050 |
0.2009083063 |
- |
- |
- |
| 18 |
Hb_000264_130 |
0.2031475685 |
- |
- |
PREDICTED: uncharacterized protein LOC103870402 [Brassica rapa] |
| 19 |
Hb_105602_020 |
0.2090531619 |
- |
- |
PREDICTED: 60S ribosomal protein L19-1-like [Populus euphratica] |
| 20 |
Hb_001433_100 |
0.2117029214 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 3 isoform X1 [Jatropha curcas] |