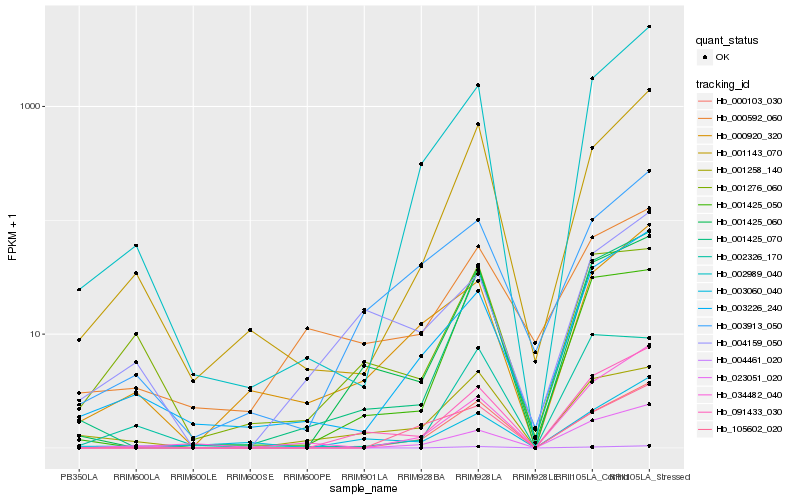
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001425_060 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_091433_030 |
0.0700268792 |
- |
- |
- |
| 3 |
Hb_001425_070 |
0.0966958418 |
- |
- |
- |
| 4 |
Hb_001425_050 |
0.1404069938 |
- |
- |
- |
| 5 |
Hb_000103_030 |
0.1485898119 |
- |
- |
hypothetical protein VITISV_021700 [Vitis vinifera] |
| 6 |
Hb_004461_020 |
0.170143327 |
- |
- |
PREDICTED: uncharacterized protein LOC104105697 [Nicotiana tomentosiformis] |
| 7 |
Hb_003060_040 |
0.1722109409 |
transcription factor |
TF Family: MIKC |
PREDICTED: agamous-like MADS-box protein AGL15 [Jatropha curcas] |
| 8 |
Hb_002989_040 |
0.1757374246 |
- |
- |
hypothetical protein 1 [Hevea brasiliensis] |
| 9 |
Hb_023051_020 |
0.176385649 |
- |
- |
- |
| 10 |
Hb_001258_140 |
0.1779723492 |
- |
- |
- |
| 11 |
Hb_001143_070 |
0.1784045702 |
- |
- |
Acyl-CoA N-acyltransferases (NAT) superfamily protein isoform 1 [Theobroma cacao] |
| 12 |
Hb_003913_050 |
0.1796034806 |
- |
- |
60S ribosomal protein L27B [Hevea brasiliensis] |
| 13 |
Hb_003226_240 |
0.189022367 |
- |
- |
PREDICTED: caffeoylshikimate esterase [Jatropha curcas] |
| 14 |
Hb_034482_040 |
0.1945975295 |
- |
- |
aspartate kinase, partial [Francisella tularensis] |
| 15 |
Hb_002326_170 |
0.1948055121 |
- |
- |
Protein Z, putative [Ricinus communis] |
| 16 |
Hb_000920_320 |
0.1981709871 |
transcription factor |
TF Family: Orphans |
hypothetical protein JCGZ_24525 [Jatropha curcas] |
| 17 |
Hb_004159_050 |
0.2008412515 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001276_060 |
0.2030284438 |
- |
- |
hypothetical protein POPTR_0015s00420g [Populus trichocarpa] |
| 19 |
Hb_000592_060 |
0.2062958847 |
- |
- |
PREDICTED: ras-related protein RABA6a [Populus euphratica] |
| 20 |
Hb_105602_020 |
0.2087713192 |
- |
- |
PREDICTED: 60S ribosomal protein L19-1-like [Populus euphratica] |