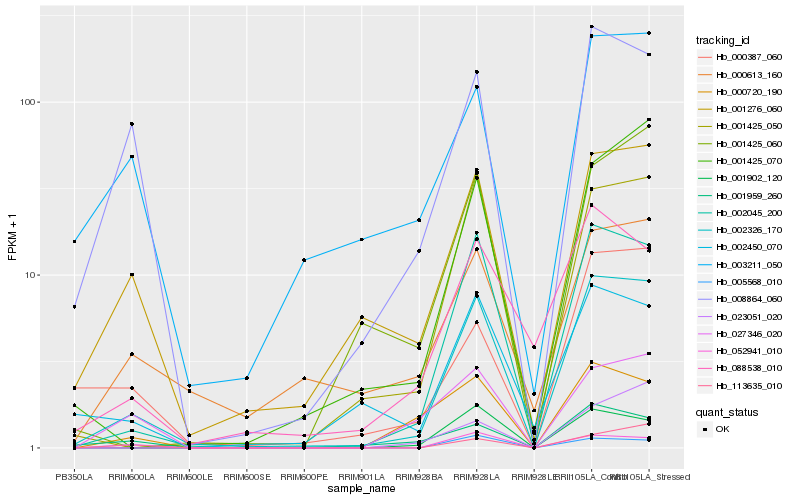
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002326_170 |
0.0 |
- |
- |
Protein Z, putative [Ricinus communis] |
| 2 |
Hb_002045_200 |
0.105590875 |
- |
- |
PREDICTED: beta-carotene isomerase D27, chloroplastic [Jatropha curcas] |
| 3 |
Hb_001425_050 |
0.1504075575 |
- |
- |
- |
| 4 |
Hb_001276_060 |
0.1555393833 |
- |
- |
hypothetical protein POPTR_0015s00420g [Populus trichocarpa] |
| 5 |
Hb_008864_060 |
0.1752432397 |
- |
- |
PREDICTED: uncharacterized protein LOC105634361 [Jatropha curcas] |
| 6 |
Hb_001425_070 |
0.1796080777 |
- |
- |
- |
| 7 |
Hb_001902_120 |
0.1831729093 |
- |
- |
PREDICTED: uncharacterized protein LOC105648455 isoform X2 [Jatropha curcas] |
| 8 |
Hb_052941_010 |
0.186241926 |
- |
- |
PREDICTED: probable peroxidase 61 [Jatropha curcas] |
| 9 |
Hb_000387_060 |
0.1902760542 |
- |
- |
- |
| 10 |
Hb_005568_010 |
0.1918281624 |
- |
- |
- |
| 11 |
Hb_023051_020 |
0.1921852626 |
- |
- |
- |
| 12 |
Hb_002450_070 |
0.193799941 |
- |
- |
(S)-N-methylcoclaurine 3'-hydroxylase isozyme, putative [Ricinus communis] |
| 13 |
Hb_113635_010 |
0.1940280084 |
- |
- |
- |
| 14 |
Hb_001425_060 |
0.1948055121 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001959_260 |
0.1969042438 |
- |
- |
- |
| 16 |
Hb_000720_190 |
0.197826318 |
- |
- |
unknown [Populus trichocarpa x Populus deltoides] |
| 17 |
Hb_027346_020 |
0.1995440487 |
- |
- |
- |
| 18 |
Hb_088538_010 |
0.2003584888 |
- |
- |
hypothetical protein JCGZ_12177 [Jatropha curcas] |
| 19 |
Hb_000613_160 |
0.2030322324 |
- |
- |
coated vesicle membrane protein, putative [Ricinus communis] |
| 20 |
Hb_003211_050 |
0.2031030108 |
- |
- |
PREDICTED: RNA-binding protein 24-B isoform X1 [Jatropha curcas] |