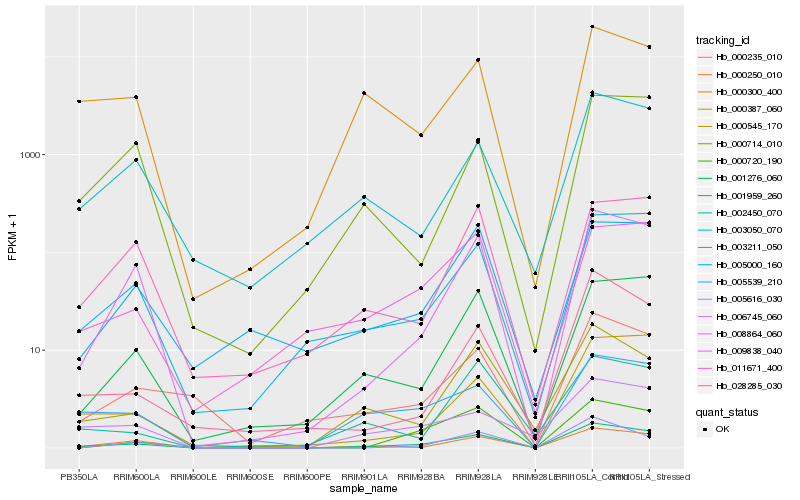
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001959_260 |
0.0 |
- |
- |
- |
| 2 |
Hb_008864_060 |
0.132660224 |
- |
- |
PREDICTED: uncharacterized protein LOC105634361 [Jatropha curcas] |
| 3 |
Hb_003211_050 |
0.1475527902 |
- |
- |
PREDICTED: RNA-binding protein 24-B isoform X1 [Jatropha curcas] |
| 4 |
Hb_000250_010 |
0.1490000227 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
| 5 |
Hb_005539_210 |
0.1582940376 |
- |
- |
- |
| 6 |
Hb_001276_060 |
0.1606998394 |
- |
- |
hypothetical protein POPTR_0015s00420g [Populus trichocarpa] |
| 7 |
Hb_000545_170 |
0.1620460258 |
- |
- |
PREDICTED: putative calcium-transporting ATPase 11, plasma membrane-type isoform X2 [Jatropha curcas] |
| 8 |
Hb_000300_400 |
0.173693582 |
- |
- |
hypothetical protein PHAVU_001G114100g [Phaseolus vulgaris] |
| 9 |
Hb_006745_060 |
0.1739650235 |
- |
- |
- |
| 10 |
Hb_003050_070 |
0.1769541749 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000387_060 |
0.1772917137 |
- |
- |
- |
| 12 |
Hb_000235_010 |
0.1815633273 |
- |
- |
PREDICTED: serine carboxypeptidase-like 18 [Jatropha curcas] |
| 13 |
Hb_000720_190 |
0.1846868659 |
- |
- |
unknown [Populus trichocarpa x Populus deltoides] |
| 14 |
Hb_009838_040 |
0.1847448026 |
- |
- |
hypothetical protein JCGZ_12866 [Jatropha curcas] |
| 15 |
Hb_028285_030 |
0.185970931 |
- |
- |
PREDICTED: protein GLUTAMINE DUMPER 1-like [Elaeis guineensis] |
| 16 |
Hb_011671_400 |
0.1907346082 |
- |
- |
PREDICTED: cyclin-U4-1 [Jatropha curcas] |
| 17 |
Hb_000714_010 |
0.1918507519 |
- |
- |
PREDICTED: salt stress-induced hydrophobic peptide ESI3 [Vitis vinifera] |
| 18 |
Hb_002450_070 |
0.1921257462 |
- |
- |
(S)-N-methylcoclaurine 3'-hydroxylase isozyme, putative [Ricinus communis] |
| 19 |
Hb_005000_160 |
0.1946944485 |
- |
- |
PREDICTED: uncharacterized protein LOC105637882 [Jatropha curcas] |
| 20 |
Hb_005616_030 |
0.1958178191 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 1 [Eucalyptus grandis] |