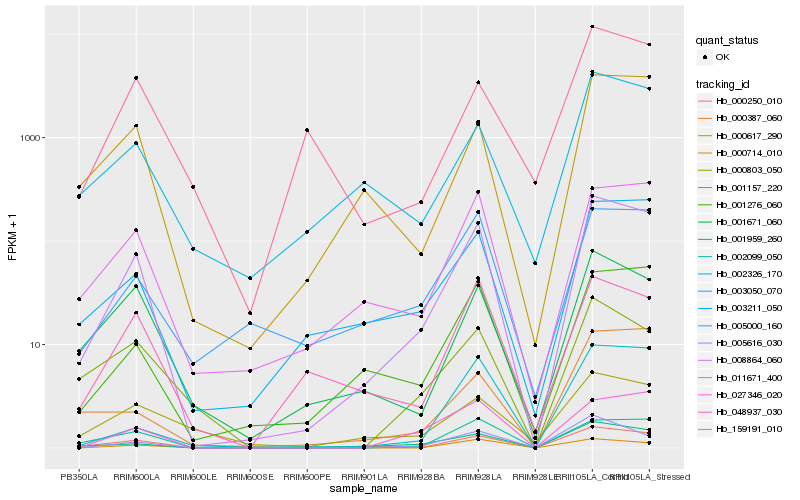
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008864_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105634361 [Jatropha curcas] |
| 2 |
Hb_000250_010 |
0.1054710537 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
| 3 |
Hb_001959_260 |
0.132660224 |
- |
- |
- |
| 4 |
Hb_011671_400 |
0.1490787614 |
- |
- |
PREDICTED: cyclin-U4-1 [Jatropha curcas] |
| 5 |
Hb_001276_060 |
0.1558540639 |
- |
- |
hypothetical protein POPTR_0015s00420g [Populus trichocarpa] |
| 6 |
Hb_002099_050 |
0.1560721048 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_003211_050 |
0.1590230459 |
- |
- |
PREDICTED: RNA-binding protein 24-B isoform X1 [Jatropha curcas] |
| 8 |
Hb_000714_010 |
0.1652290097 |
- |
- |
PREDICTED: salt stress-induced hydrophobic peptide ESI3 [Vitis vinifera] |
| 9 |
Hb_001671_060 |
0.1711353318 |
- |
- |
fad NAD binding oxidoreductases, putative [Ricinus communis] |
| 10 |
Hb_005616_030 |
0.172994905 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 1 [Eucalyptus grandis] |
| 11 |
Hb_002326_170 |
0.1752432397 |
- |
- |
Protein Z, putative [Ricinus communis] |
| 12 |
Hb_000617_290 |
0.1778738194 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_027346_020 |
0.1802350686 |
- |
- |
- |
| 14 |
Hb_003050_070 |
0.1856139564 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_048937_030 |
0.1869922317 |
- |
- |
hypothetical protein RCOM_0231790 [Ricinus communis] |
| 16 |
Hb_005000_160 |
0.1911245935 |
- |
- |
PREDICTED: uncharacterized protein LOC105637882 [Jatropha curcas] |
| 17 |
Hb_001157_220 |
0.1913466772 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000387_060 |
0.1955598405 |
- |
- |
- |
| 19 |
Hb_000803_050 |
0.1966215906 |
- |
- |
PREDICTED: uncharacterized protein LOC105648296 [Jatropha curcas] |
| 20 |
Hb_159191_010 |
0.2053124624 |
- |
- |
protease inhibitor protein [Hevea brasiliensis] |