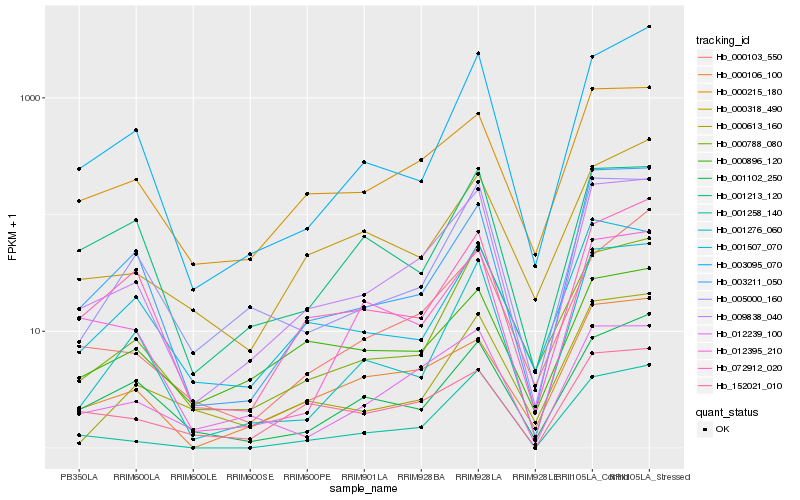
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009838_040 |
0.0 |
- |
- |
hypothetical protein JCGZ_12866 [Jatropha curcas] |
| 2 |
Hb_000215_180 |
0.1061401842 |
- |
- |
hypothetical protein 17 [Hevea brasiliensis] |
| 3 |
Hb_012239_100 |
0.1171311428 |
- |
- |
hypothetical protein VITISV_043897 [Vitis vinifera] |
| 4 |
Hb_005000_160 |
0.1179754529 |
- |
- |
PREDICTED: uncharacterized protein LOC105637882 [Jatropha curcas] |
| 5 |
Hb_000788_080 |
0.1232649062 |
- |
- |
PREDICTED: uncharacterized protein LOC105629703 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001258_140 |
0.1287178194 |
- |
- |
- |
| 7 |
Hb_000106_100 |
0.1340551013 |
- |
- |
- |
| 8 |
Hb_003211_050 |
0.1356523252 |
- |
- |
PREDICTED: RNA-binding protein 24-B isoform X1 [Jatropha curcas] |
| 9 |
Hb_012395_210 |
0.1380655507 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001276_060 |
0.1388811911 |
- |
- |
hypothetical protein POPTR_0015s00420g [Populus trichocarpa] |
| 11 |
Hb_003095_070 |
0.1406708945 |
- |
- |
hypothetical protein POPTR_0009s11230g [Populus trichocarpa] |
| 12 |
Hb_072912_020 |
0.1425293095 |
- |
- |
abhydrolase domain containing, putative [Ricinus communis] |
| 13 |
Hb_001507_070 |
0.1436378567 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001102_250 |
0.1460791591 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase CCNB1IP1 homolog isoform X1 [Jatropha curcas] |
| 15 |
Hb_000896_120 |
0.1503198401 |
- |
- |
PREDICTED: acyl-protein thioesterase 2-like [Populus euphratica] |
| 16 |
Hb_001213_120 |
0.1509082871 |
- |
- |
PREDICTED: immediate early response 3-interacting protein 1-like [Brassica rapa] |
| 17 |
Hb_152021_010 |
0.1519621972 |
- |
- |
PREDICTED: uncharacterized protein LOC105639312 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000613_160 |
0.1538769609 |
- |
- |
coated vesicle membrane protein, putative [Ricinus communis] |
| 19 |
Hb_000318_490 |
0.1544988308 |
- |
- |
PREDICTED: mitochondrial import receptor subunit TOM6 homolog [Jatropha curcas] |
| 20 |
Hb_000103_550 |
0.1581946057 |
- |
- |
PREDICTED: guanine nucleotide-binding protein subunit gamma 2 isoform X1 [Jatropha curcas] |