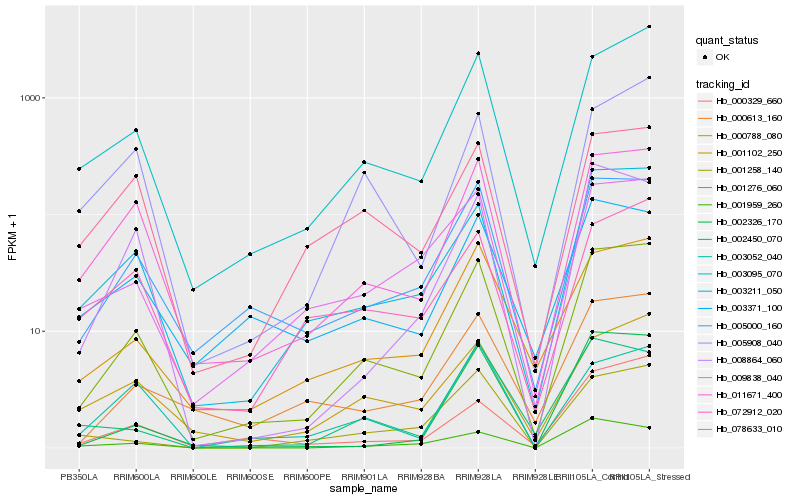
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001276_060 |
0.0 |
- |
- |
hypothetical protein POPTR_0015s00420g [Populus trichocarpa] |
| 2 |
Hb_003095_070 |
0.1018368428 |
- |
- |
hypothetical protein POPTR_0009s11230g [Populus trichocarpa] |
| 3 |
Hb_011671_400 |
0.1051062283 |
- |
- |
PREDICTED: cyclin-U4-1 [Jatropha curcas] |
| 4 |
Hb_003211_050 |
0.1076880201 |
- |
- |
PREDICTED: RNA-binding protein 24-B isoform X1 [Jatropha curcas] |
| 5 |
Hb_005000_160 |
0.114113058 |
- |
- |
PREDICTED: uncharacterized protein LOC105637882 [Jatropha curcas] |
| 6 |
Hb_000788_080 |
0.1288062744 |
- |
- |
PREDICTED: uncharacterized protein LOC105629703 isoform X1 [Jatropha curcas] |
| 7 |
Hb_009838_040 |
0.1388811911 |
- |
- |
hypothetical protein JCGZ_12866 [Jatropha curcas] |
| 8 |
Hb_000613_160 |
0.1430549152 |
- |
- |
coated vesicle membrane protein, putative [Ricinus communis] |
| 9 |
Hb_001102_250 |
0.1459506504 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase CCNB1IP1 homolog isoform X1 [Jatropha curcas] |
| 10 |
Hb_005908_040 |
0.1528273914 |
- |
- |
hypothetical protein POPTR_0017s06850g [Populus trichocarpa] |
| 11 |
Hb_002326_170 |
0.1555393833 |
- |
- |
Protein Z, putative [Ricinus communis] |
| 12 |
Hb_008864_060 |
0.1558540639 |
- |
- |
PREDICTED: uncharacterized protein LOC105634361 [Jatropha curcas] |
| 13 |
Hb_000329_660 |
0.1570174521 |
- |
- |
PREDICTED: carotenoid cleavage dioxygenase 7, chloroplastic [Jatropha curcas] |
| 14 |
Hb_078633_010 |
0.1571300931 |
- |
- |
PREDICTED: RNA-binding protein 24-B isoform X1 [Jatropha curcas] |
| 15 |
Hb_002450_070 |
0.1579207255 |
- |
- |
(S)-N-methylcoclaurine 3'-hydroxylase isozyme, putative [Ricinus communis] |
| 16 |
Hb_001959_260 |
0.1606998394 |
- |
- |
- |
| 17 |
Hb_001258_140 |
0.1631947437 |
- |
- |
- |
| 18 |
Hb_072912_020 |
0.1635549696 |
- |
- |
abhydrolase domain containing, putative [Ricinus communis] |
| 19 |
Hb_003052_040 |
0.1663325195 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_003371_100 |
0.1663350778 |
transcription factor |
TF Family: HB |
homeobox protein, putative [Ricinus communis] |