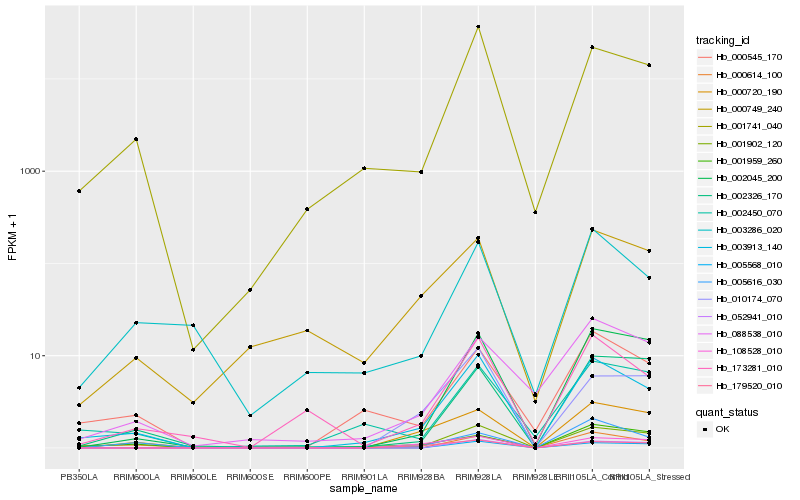
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003913_140 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105640324 [Jatropha curcas] |
| 2 |
Hb_001741_040 |
0.1310657661 |
rubber biosynthesis |
Gene Name: REF3 |
REF-like stress related protein 1 [Hevea brasiliensis] |
| 3 |
Hb_001902_120 |
0.1637326614 |
- |
- |
PREDICTED: uncharacterized protein LOC105648455 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000545_170 |
0.1759240657 |
- |
- |
PREDICTED: putative calcium-transporting ATPase 11, plasma membrane-type isoform X2 [Jatropha curcas] |
| 5 |
Hb_173281_010 |
0.1780441411 |
- |
- |
Pectin lyase-like superfamily protein isoform 1 [Theobroma cacao] |
| 6 |
Hb_002045_200 |
0.1834629429 |
- |
- |
PREDICTED: beta-carotene isomerase D27, chloroplastic [Jatropha curcas] |
| 7 |
Hb_002450_070 |
0.1930710285 |
- |
- |
(S)-N-methylcoclaurine 3'-hydroxylase isozyme, putative [Ricinus communis] |
| 8 |
Hb_000720_190 |
0.1966577717 |
- |
- |
unknown [Populus trichocarpa x Populus deltoides] |
| 9 |
Hb_003286_020 |
0.197438911 |
- |
- |
hypothetical protein POPTR_0007s05070g [Populus trichocarpa] |
| 10 |
Hb_179520_010 |
0.2103542933 |
- |
- |
PREDICTED: uncharacterized protein LOC105763691 [Gossypium raimondii] |
| 11 |
Hb_005568_010 |
0.212539819 |
- |
- |
- |
| 12 |
Hb_052941_010 |
0.2132843996 |
- |
- |
PREDICTED: probable peroxidase 61 [Jatropha curcas] |
| 13 |
Hb_000749_240 |
0.214565337 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_010174_070 |
0.2168558843 |
- |
- |
- |
| 15 |
Hb_088538_010 |
0.2193525572 |
- |
- |
hypothetical protein JCGZ_12177 [Jatropha curcas] |
| 16 |
Hb_001959_260 |
0.2201637424 |
- |
- |
- |
| 17 |
Hb_108528_010 |
0.2204297132 |
- |
- |
unnamed protein product [Coffea canephora] |
| 18 |
Hb_002326_170 |
0.2210765368 |
- |
- |
Protein Z, putative [Ricinus communis] |
| 19 |
Hb_005616_030 |
0.2211120001 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 1 [Eucalyptus grandis] |
| 20 |
Hb_000614_100 |
0.2219280686 |
- |
- |
3-oxoacyl-[acyl-carrier-protein] reductase [Populus trichocarpa] |