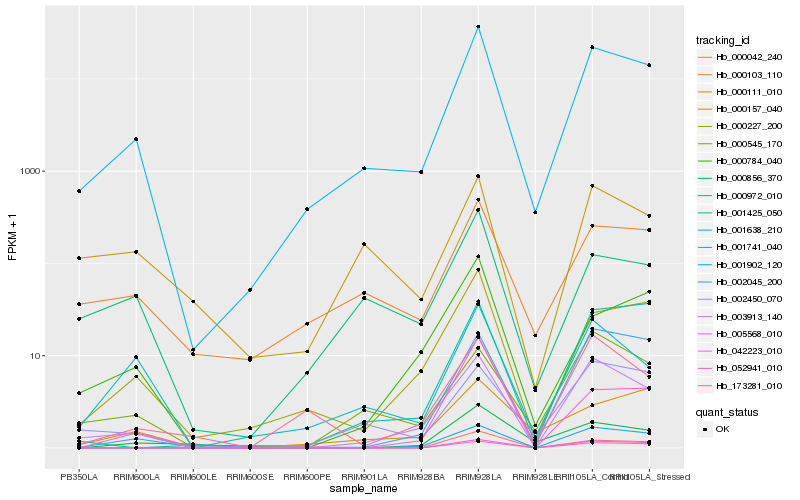
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001741_040 |
0.0 |
rubber biosynthesis |
Gene Name: REF3 |
REF-like stress related protein 1 [Hevea brasiliensis] |
| 2 |
Hb_003913_140 |
0.1310657661 |
- |
- |
PREDICTED: uncharacterized protein LOC105640324 [Jatropha curcas] |
| 3 |
Hb_000227_200 |
0.14860948 |
- |
- |
PREDICTED: protein FANTASTIC FOUR 3 [Populus euphratica] |
| 4 |
Hb_000103_110 |
0.1594964805 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_002450_070 |
0.1709129849 |
- |
- |
(S)-N-methylcoclaurine 3'-hydroxylase isozyme, putative [Ricinus communis] |
| 6 |
Hb_001638_210 |
0.1857954402 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000972_010 |
0.191329301 |
- |
- |
PREDICTED: kinesin KP1 [Jatropha curcas] |
| 8 |
Hb_001902_120 |
0.1943980958 |
- |
- |
PREDICTED: uncharacterized protein LOC105648455 isoform X2 [Jatropha curcas] |
| 9 |
Hb_173281_010 |
0.1957699802 |
- |
- |
Pectin lyase-like superfamily protein isoform 1 [Theobroma cacao] |
| 10 |
Hb_002045_200 |
0.1991300797 |
- |
- |
PREDICTED: beta-carotene isomerase D27, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000545_170 |
0.1998170359 |
- |
- |
PREDICTED: putative calcium-transporting ATPase 11, plasma membrane-type isoform X2 [Jatropha curcas] |
| 12 |
Hb_000111_010 |
0.2004630933 |
- |
- |
DNA-directed RNA polymerase, putative [Ricinus communis] |
| 13 |
Hb_000856_370 |
0.2025042672 |
- |
- |
PREDICTED: protein NETWORKED 1A [Jatropha curcas] |
| 14 |
Hb_000784_040 |
0.2038095493 |
- |
- |
hypothetical protein POPTR_0013s11550g [Populus trichocarpa] |
| 15 |
Hb_000042_240 |
0.2048181472 |
- |
- |
- |
| 16 |
Hb_001425_050 |
0.2048966758 |
- |
- |
- |
| 17 |
Hb_005568_010 |
0.2060358961 |
- |
- |
- |
| 18 |
Hb_052941_010 |
0.208898892 |
- |
- |
PREDICTED: probable peroxidase 61 [Jatropha curcas] |
| 19 |
Hb_000157_040 |
0.2097056861 |
- |
- |
PREDICTED: cycloartenol-C-24-methyltransferase [Jatropha curcas] |
| 20 |
Hb_042223_010 |
0.2105771578 |
- |
- |
Protein Z, putative [Ricinus communis] |