| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
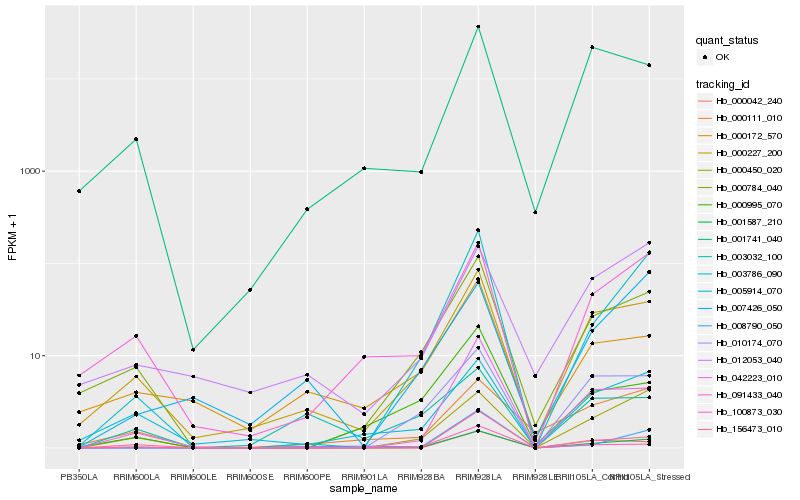
Hb_001587_210 |
0.0 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At2g19130 [Jatropha curcas] |
| 2 |
Hb_000227_200 |
0.1477619515 |
- |
- |
PREDICTED: protein FANTASTIC FOUR 3 [Populus euphratica] |
| 3 |
Hb_042223_010 |
0.1782198072 |
- |
- |
Protein Z, putative [Ricinus communis] |
| 4 |
Hb_000784_040 |
0.1934741602 |
- |
- |
hypothetical protein POPTR_0013s11550g [Populus trichocarpa] |
| 5 |
Hb_003786_090 |
0.1941870185 |
- |
- |
hypothetical protein JCGZ_19199 [Jatropha curcas] |
| 6 |
Hb_091433_040 |
0.2122561439 |
- |
- |
PREDICTED: josephin-like protein [Jatropha curcas] |
| 7 |
Hb_000450_020 |
0.2196304231 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_156473_010 |
0.2205346777 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Prunus mume] |
| 9 |
Hb_000172_570 |
0.2212388938 |
- |
- |
PREDICTED: uncharacterized protein LOC105636518 isoform X1 [Jatropha curcas] |
| 10 |
Hb_008790_050 |
0.2234572903 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_003032_100 |
0.2284798207 |
- |
- |
PREDICTED: GDP-mannose transporter GONST4 [Jatropha curcas] |
| 12 |
Hb_010174_070 |
0.2323147059 |
- |
- |
- |
| 13 |
Hb_001741_040 |
0.2335413481 |
rubber biosynthesis |
Gene Name: REF3 |
REF-like stress related protein 1 [Hevea brasiliensis] |
| 14 |
Hb_012053_040 |
0.2336688812 |
- |
- |
PREDICTED: putative phospholipid:diacylglycerol acyltransferase 2 [Jatropha curcas] |
| 15 |
Hb_005914_070 |
0.2347690815 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_007426_050 |
0.2352950727 |
transcription factor |
TF Family: Tify |
JAZ8 [Hevea brasiliensis] |
| 17 |
Hb_000042_240 |
0.2358429061 |
- |
- |
- |
| 18 |
Hb_000111_010 |
0.2443320833 |
- |
- |
DNA-directed RNA polymerase, putative [Ricinus communis] |
| 19 |
Hb_000995_070 |
0.2588427469 |
- |
- |
PREDICTED: secoisolariciresinol dehydrogenase-like [Jatropha curcas] |
| 20 |
Hb_100873_030 |
0.2595143794 |
- |
- |
PREDICTED: uncharacterized protein LOC105795653 [Gossypium raimondii] |