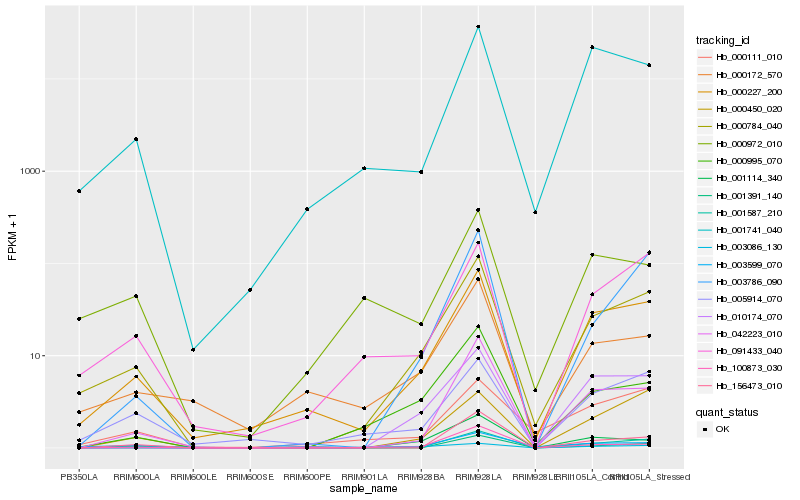
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000784_040 |
0.0 |
- |
- |
hypothetical protein POPTR_0013s11550g [Populus trichocarpa] |
| 2 |
Hb_000227_200 |
0.1176892532 |
- |
- |
PREDICTED: protein FANTASTIC FOUR 3 [Populus euphratica] |
| 3 |
Hb_091433_040 |
0.163232007 |
- |
- |
PREDICTED: josephin-like protein [Jatropha curcas] |
| 4 |
Hb_000995_070 |
0.1717320644 |
- |
- |
PREDICTED: secoisolariciresinol dehydrogenase-like [Jatropha curcas] |
| 5 |
Hb_042223_010 |
0.1796633504 |
- |
- |
Protein Z, putative [Ricinus communis] |
| 6 |
Hb_005914_070 |
0.1864591128 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_003599_070 |
0.1901434403 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 8 |
Hb_003786_090 |
0.1916520458 |
- |
- |
hypothetical protein JCGZ_19199 [Jatropha curcas] |
| 9 |
Hb_001587_210 |
0.1934741602 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At2g19130 [Jatropha curcas] |
| 10 |
Hb_000172_570 |
0.1944694887 |
- |
- |
PREDICTED: uncharacterized protein LOC105636518 isoform X1 [Jatropha curcas] |
| 11 |
Hb_010174_070 |
0.1960209577 |
- |
- |
- |
| 12 |
Hb_001741_040 |
0.2038095493 |
rubber biosynthesis |
Gene Name: REF3 |
REF-like stress related protein 1 [Hevea brasiliensis] |
| 13 |
Hb_000450_020 |
0.2044817941 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000111_010 |
0.2060028948 |
- |
- |
DNA-directed RNA polymerase, putative [Ricinus communis] |
| 15 |
Hb_000972_010 |
0.2138614795 |
- |
- |
PREDICTED: kinesin KP1 [Jatropha curcas] |
| 16 |
Hb_100873_030 |
0.2140021138 |
- |
- |
PREDICTED: uncharacterized protein LOC105795653 [Gossypium raimondii] |
| 17 |
Hb_003086_130 |
0.2189161005 |
- |
- |
PREDICTED: uncharacterized protein LOC105631113 [Jatropha curcas] |
| 18 |
Hb_156473_010 |
0.219577233 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Prunus mume] |
| 19 |
Hb_001114_340 |
0.2254384952 |
- |
- |
hypothetical protein JCGZ_13805 [Jatropha curcas] |
| 20 |
Hb_001391_140 |
0.2336373728 |
- |
- |
PREDICTED: uncharacterized protein LOC101293660 [Fragaria vesca subsp. vesca] |