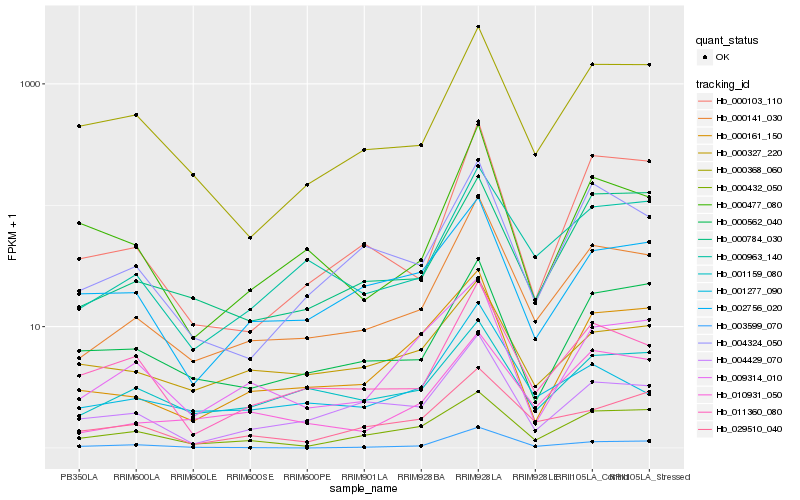
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000141_030 |
0.0 |
- |
- |
PREDICTED: scarecrow-like protein 22 [Jatropha curcas] |
| 2 |
Hb_000963_140 |
0.1163197814 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 3 |
Hb_010931_050 |
0.1312901477 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ25-like [Jatropha curcas] |
| 4 |
Hb_011360_080 |
0.1365640703 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 5 |
Hb_004429_070 |
0.1373794615 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase RPK2 [Jatropha curcas] |
| 6 |
Hb_004324_050 |
0.1388731904 |
- |
- |
PREDICTED: spermine synthase [Jatropha curcas] |
| 7 |
Hb_000368_060 |
0.1423201665 |
- |
- |
thioredoxin H-type 6 [Hevea brasiliensis] |
| 8 |
Hb_000103_110 |
0.1459508846 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_001159_080 |
0.1466117663 |
- |
- |
hypothetical protein JCGZ_20745 [Jatropha curcas] |
| 10 |
Hb_000161_150 |
0.1467314563 |
- |
- |
PREDICTED: protein argonaute 2-like [Jatropha curcas] |
| 11 |
Hb_000784_030 |
0.1468100697 |
- |
- |
neutral/alkaline invertase 2 [Hevea brasiliensis] |
| 12 |
Hb_000562_040 |
0.1518509123 |
- |
- |
PREDICTED: auxin response factor 5 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000477_080 |
0.1530034443 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 11 [Jatropha curcas] |
| 14 |
Hb_000432_050 |
0.1536222164 |
- |
- |
GTP-binding protein typa/bipa, putative [Ricinus communis] |
| 15 |
Hb_029510_040 |
0.1576097226 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g23330 [Vitis vinifera] |
| 16 |
Hb_002756_020 |
0.1590489367 |
transcription factor |
TF Family: HB |
bel1 homeotic protein, putative [Ricinus communis] |
| 17 |
Hb_001277_090 |
0.1613341042 |
- |
- |
PREDICTED: uncharacterized protein LOC105632061 [Jatropha curcas] |
| 18 |
Hb_009314_010 |
0.1616880127 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 19 |
Hb_003599_070 |
0.1653941357 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 20 |
Hb_000327_220 |
0.166892947 |
- |
- |
PREDICTED: probable protein phosphatase 2C 38 [Jatropha curcas] |