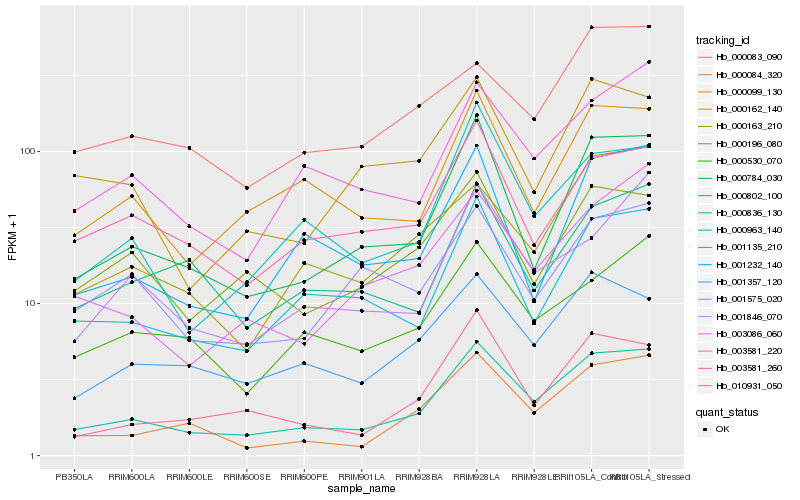
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000836_130 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105642953 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000784_030 |
0.1017230963 |
- |
- |
neutral/alkaline invertase 2 [Hevea brasiliensis] |
| 3 |
Hb_000084_320 |
0.1084919626 |
- |
- |
PREDICTED: probable small nuclear ribonucleoprotein G [Populus euphratica] |
| 4 |
Hb_001135_210 |
0.1173006236 |
- |
- |
PREDICTED: probable adenylate kinase 6, chloroplastic [Jatropha curcas] |
| 5 |
Hb_001357_120 |
0.1178616737 |
- |
- |
ubiquitin-activating enzyme E1, putative [Ricinus communis] |
| 6 |
Hb_003581_220 |
0.1182482292 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 7 |
Hb_000099_130 |
0.1196275182 |
- |
- |
PREDICTED: uncharacterized protein LOC105645728 [Jatropha curcas] |
| 8 |
Hb_010931_050 |
0.120889912 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ25-like [Jatropha curcas] |
| 9 |
Hb_000963_140 |
0.1261721289 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 10 |
Hb_000530_070 |
0.1271233951 |
- |
- |
neutral/alkaline invertase 3 [Hevea brasiliensis] |
| 11 |
Hb_003581_260 |
0.127447264 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 29-like [Jatropha curcas] |
| 12 |
Hb_000163_210 |
0.1302695953 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 5-B [Tarenaya hassleriana] |
| 13 |
Hb_001232_140 |
0.1318374471 |
- |
- |
PREDICTED: uncharacterized protein LOC105634966 [Jatropha curcas] |
| 14 |
Hb_003086_060 |
0.1318955297 |
- |
- |
PREDICTED: uncharacterized protein LOC105629937 [Jatropha curcas] |
| 15 |
Hb_000196_080 |
0.1319640679 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH79-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_000162_140 |
0.1373125019 |
transcription factor |
TF Family: CSD |
PREDICTED: glycine-rich protein 2 [Jatropha curcas] |
| 17 |
Hb_000802_100 |
0.1376267953 |
- |
- |
PREDICTED: uncharacterized protein LOC105634009 [Jatropha curcas] |
| 18 |
Hb_000083_090 |
0.1397083313 |
- |
- |
immunophilin, putative [Ricinus communis] |
| 19 |
Hb_001575_020 |
0.1400715506 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001846_070 |
0.1408647087 |
- |
- |
PREDICTED: fructokinase-1 [Jatropha curcas] |