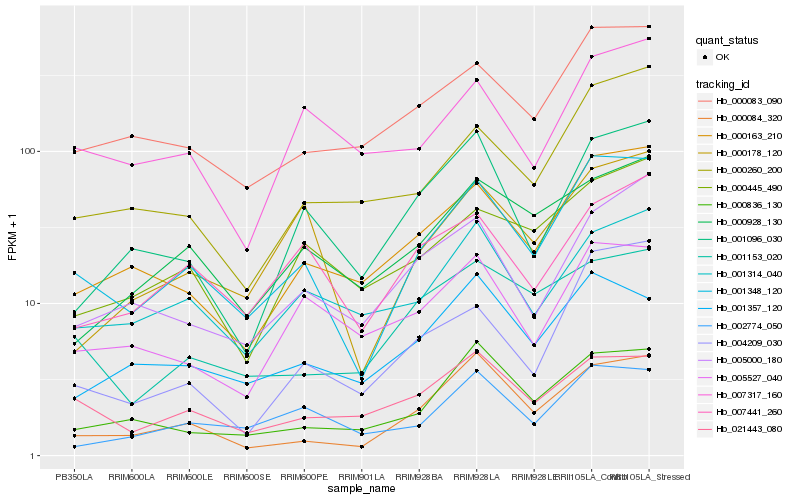
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001348_120 |
0.0 |
- |
- |
Phloem protein 2-B10, putative [Theobroma cacao] |
| 2 |
Hb_000163_210 |
0.126114472 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 5-B [Tarenaya hassleriana] |
| 3 |
Hb_000083_090 |
0.1312639281 |
- |
- |
immunophilin, putative [Ricinus communis] |
| 4 |
Hb_000084_320 |
0.136198726 |
- |
- |
PREDICTED: probable small nuclear ribonucleoprotein G [Populus euphratica] |
| 5 |
Hb_000260_200 |
0.1482589378 |
- |
- |
- |
| 6 |
Hb_000836_130 |
0.148898175 |
- |
- |
PREDICTED: uncharacterized protein LOC105642953 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000178_120 |
0.1496771729 |
- |
- |
PREDICTED: histone H2B [Jatropha curcas] |
| 8 |
Hb_002774_050 |
0.1511934648 |
- |
- |
PREDICTED: uncharacterized protein LOC105630601 [Jatropha curcas] |
| 9 |
Hb_004209_030 |
0.151346484 |
- |
- |
PREDICTED: (DL)-glycerol-3-phosphatase 2 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001314_040 |
0.1523357184 |
- |
- |
hypothetical protein POPTR_0011s10470g [Populus trichocarpa] |
| 11 |
Hb_007441_260 |
0.1540278639 |
- |
- |
PREDICTED: ATP synthase subunit delta', mitochondrial-like [Jatropha curcas] |
| 12 |
Hb_005000_180 |
0.1554739953 |
- |
- |
hypothetical protein JCGZ_07408 [Jatropha curcas] |
| 13 |
Hb_000445_490 |
0.1574603638 |
- |
- |
- |
| 14 |
Hb_001096_030 |
0.1584726684 |
- |
- |
uridylate kinase plant, putative [Ricinus communis] |
| 15 |
Hb_001153_020 |
0.1592457699 |
- |
- |
PREDICTED: histone deacetylase complex subunit SAP18-like [Cucumis melo] |
| 16 |
Hb_021443_080 |
0.159506113 |
- |
- |
PREDICTED: CDGSH iron-sulfur domain-containing protein NEET [Jatropha curcas] |
| 17 |
Hb_001357_120 |
0.1607460886 |
- |
- |
ubiquitin-activating enzyme E1, putative [Ricinus communis] |
| 18 |
Hb_000928_130 |
0.1608465951 |
- |
- |
PREDICTED: protein BOLA4, chloroplastic/mitochondrial [Jatropha curcas] |
| 19 |
Hb_005527_040 |
0.1618053554 |
- |
- |
PREDICTED: methionine adenosyltransferase 2 subunit beta [Jatropha curcas] |
| 20 |
Hb_007317_160 |
0.1640141205 |
- |
- |
PREDICTED: uncharacterized protein At2g27730, mitochondrial [Jatropha curcas] |