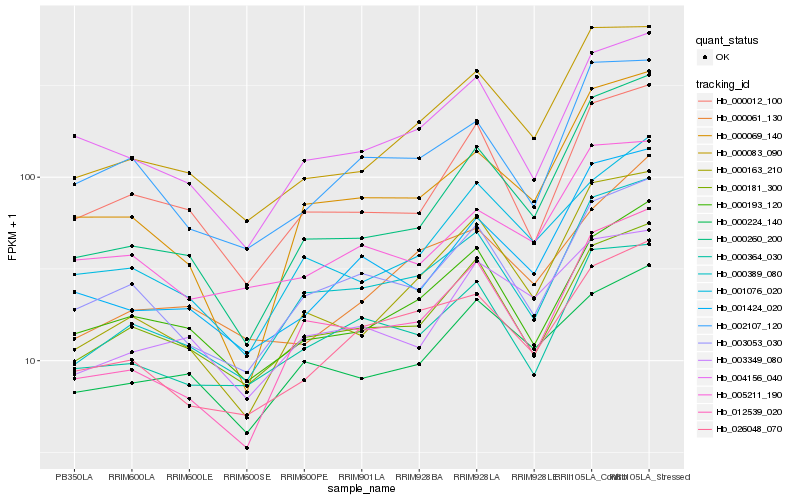
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000083_090 |
0.0 |
- |
- |
immunophilin, putative [Ricinus communis] |
| 2 |
Hb_000163_210 |
0.0666291763 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 5-B [Tarenaya hassleriana] |
| 3 |
Hb_000260_200 |
0.0930034596 |
- |
- |
- |
| 4 |
Hb_001424_020 |
0.0934052237 |
- |
- |
PREDICTED: AP-1 complex subunit sigma-2 isoform X2 [Solanum lycopersicum] |
| 5 |
Hb_000389_080 |
0.0937303983 |
- |
- |
PREDICTED: uncharacterized protein At4g26485-like [Jatropha curcas] |
| 6 |
Hb_000193_120 |
0.0963240006 |
- |
- |
PREDICTED: protein preY, mitochondrial [Jatropha curcas] |
| 7 |
Hb_004156_040 |
0.09715376 |
- |
- |
PREDICTED: uncharacterized protein LOC105110611 [Populus euphratica] |
| 8 |
Hb_000181_300 |
0.0980745727 |
- |
- |
PREDICTED: uncharacterized protein LOC105650445 isoform X2 [Jatropha curcas] |
| 9 |
Hb_000012_100 |
0.0988717337 |
- |
- |
UPF0587 protein C1orf123 homolog [Jatropha curcas] |
| 10 |
Hb_002107_120 |
0.0990180296 |
- |
- |
PREDICTED: transcription elongation factor 1 homolog [Jatropha curcas] |
| 11 |
Hb_005211_190 |
0.1005882906 |
- |
- |
PREDICTED: uncharacterized protein LOC105641994 [Jatropha curcas] |
| 12 |
Hb_003349_080 |
0.1073152059 |
- |
- |
PREDICTED: uncharacterized protein LOC105628287 [Jatropha curcas] |
| 13 |
Hb_001076_020 |
0.1075903871 |
- |
- |
PREDICTED: cytochrome b-c1 complex subunit 8 [Eucalyptus grandis] |
| 14 |
Hb_003053_030 |
0.1089756326 |
- |
- |
PREDICTED: transport and Golgi organization 2 homolog [Jatropha curcas] |
| 15 |
Hb_026048_070 |
0.1097852362 |
- |
- |
PREDICTED: uncharacterized protein LOC105645736 [Jatropha curcas] |
| 16 |
Hb_000224_140 |
0.1101001363 |
- |
- |
ribonuclease z, chloroplast, putative [Ricinus communis] |
| 17 |
Hb_012539_020 |
0.110690243 |
- |
- |
protein binding/ubiquitin-protein ligase 4 [Hevea brasiliensis] |
| 18 |
Hb_000364_030 |
0.1117246589 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 19 |
Hb_000069_140 |
0.1121234425 |
- |
- |
PREDICTED: uncharacterized protein LOC105642831 [Jatropha curcas] |
| 20 |
Hb_000061_130 |
0.1125513943 |
- |
- |
rotamase, putative [Ricinus communis] |