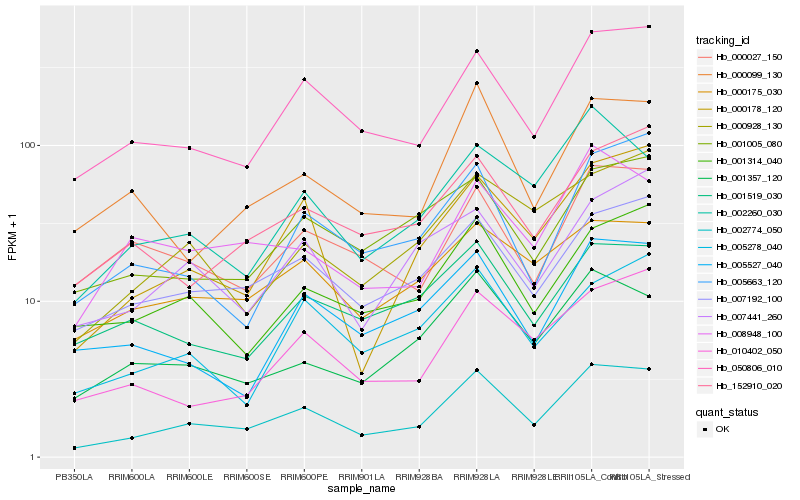
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002774_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105630601 [Jatropha curcas] |
| 2 |
Hb_050806_010 |
0.098370349 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2-17 kDa [Jatropha curcas] |
| 3 |
Hb_007192_100 |
0.1066055387 |
- |
- |
PREDICTED: protein DEHYDRATION-INDUCED 19 homolog 6-like [Jatropha curcas] |
| 4 |
Hb_001314_040 |
0.121660649 |
- |
- |
hypothetical protein POPTR_0011s10470g [Populus trichocarpa] |
| 5 |
Hb_000178_120 |
0.1233523297 |
- |
- |
PREDICTED: histone H2B [Jatropha curcas] |
| 6 |
Hb_001357_120 |
0.1256089688 |
- |
- |
ubiquitin-activating enzyme E1, putative [Ricinus communis] |
| 7 |
Hb_010402_050 |
0.1268870586 |
- |
- |
- |
| 8 |
Hb_152910_020 |
0.1274101769 |
- |
- |
PREDICTED: lactoylglutathione lyase-like [Jatropha curcas] |
| 9 |
Hb_000099_130 |
0.1295284441 |
- |
- |
PREDICTED: uncharacterized protein LOC105645728 [Jatropha curcas] |
| 10 |
Hb_000175_030 |
0.129990103 |
- |
- |
Signal recognition particle, SRP54 subunit protein [Theobroma cacao] |
| 11 |
Hb_001005_080 |
0.1304765673 |
- |
- |
PREDICTED: cytochrome b-c1 complex subunit Rieske-4, mitochondrial-like [Jatropha curcas] |
| 12 |
Hb_005663_120 |
0.1324970869 |
- |
- |
PREDICTED: uncharacterized protein LOC105635265 [Jatropha curcas] |
| 13 |
Hb_000928_130 |
0.1325335985 |
- |
- |
PREDICTED: protein BOLA4, chloroplastic/mitochondrial [Jatropha curcas] |
| 14 |
Hb_005278_040 |
0.1347182025 |
- |
- |
PREDICTED: sorcin-like isoform X1 [Populus euphratica] |
| 15 |
Hb_005527_040 |
0.1353243798 |
- |
- |
PREDICTED: methionine adenosyltransferase 2 subunit beta [Jatropha curcas] |
| 16 |
Hb_008948_100 |
0.1363167488 |
- |
- |
PREDICTED: SPX domain-containing protein 1-like [Jatropha curcas] |
| 17 |
Hb_002260_030 |
0.1364422636 |
- |
- |
PREDICTED: putative glutamine amidotransferase YLR126C [Jatropha curcas] |
| 18 |
Hb_007441_260 |
0.1371403309 |
- |
- |
PREDICTED: ATP synthase subunit delta', mitochondrial-like [Jatropha curcas] |
| 19 |
Hb_000027_150 |
0.1375991367 |
- |
- |
mitochondrial carrier protein, putative [Ricinus communis] |
| 20 |
Hb_001519_030 |
0.1408918126 |
- |
- |
PREDICTED: protein trichome birefringence-like 12 isoform X1 [Jatropha curcas] |