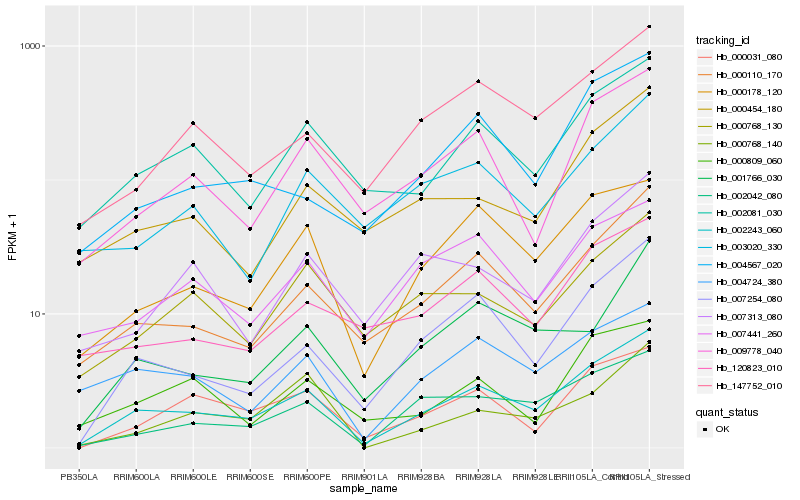
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002243_060 |
0.0 |
- |
- |
hypothetical protein POPTR_0022s00750g [Populus trichocarpa] |
| 2 |
Hb_007254_080 |
0.1240551685 |
- |
- |
- |
| 3 |
Hb_000110_170 |
0.1278990871 |
- |
- |
PREDICTED: arginine biosynthesis bifunctional protein ArgJ, chloroplastic isoform X1 [Jatropha curcas] |
| 4 |
Hb_002081_030 |
0.1386311462 |
- |
- |
unnamed protein product [Coffea canephora] |
| 5 |
Hb_002042_080 |
0.1387413375 |
rubber biosynthesis |
Gene Name: Mevalonate kinase |
mevalonate kinase [Hevea brasiliensis] |
| 6 |
Hb_000178_120 |
0.1409752165 |
- |
- |
PREDICTED: histone H2B [Jatropha curcas] |
| 7 |
Hb_147752_010 |
0.1466325621 |
- |
- |
PREDICTED: 40S ribosomal protein S9-2-like isoform X1 [Populus euphratica] |
| 8 |
Hb_000031_080 |
0.1474903859 |
- |
- |
PREDICTED: uncharacterized protein LOC105637482 [Jatropha curcas] |
| 9 |
Hb_009778_040 |
0.1480504196 |
- |
- |
PREDICTED: uncharacterized protein LOC105110611 [Populus euphratica] |
| 10 |
Hb_004567_020 |
0.1589040235 |
- |
- |
Small ubiquitin-related modifier 2 [Morus notabilis] |
| 11 |
Hb_001766_030 |
0.1609771958 |
- |
- |
hypothetical protein POPTR_0017s12640g [Populus trichocarpa] |
| 12 |
Hb_004724_380 |
0.1622454277 |
- |
- |
PREDICTED: cold-inducible RNA-binding protein B-like [Jatropha curcas] |
| 13 |
Hb_000454_180 |
0.1632444791 |
- |
- |
PREDICTED: uncharacterized protein LOC105134496 [Populus euphratica] |
| 14 |
Hb_000768_130 |
0.1663027912 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
PREDICTED: protein TRI1 [Jatropha curcas] |
| 15 |
Hb_120823_010 |
0.1669142098 |
- |
- |
- |
| 16 |
Hb_000809_060 |
0.1688179949 |
transcription factor |
TF Family: LOB |
LOB domain-containing 36 -like protein [Gossypium arboreum] |
| 17 |
Hb_007441_260 |
0.1689240722 |
- |
- |
PREDICTED: ATP synthase subunit delta', mitochondrial-like [Jatropha curcas] |
| 18 |
Hb_007313_080 |
0.1692497426 |
- |
- |
hypothetical protein CICLE_v10002804mg [Citrus clementina] |
| 19 |
Hb_003020_330 |
0.1724566511 |
- |
- |
hypothetical protein B456_010G173100 [Gossypium raimondii] |
| 20 |
Hb_000768_140 |
0.1743739839 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
tRNA (guanine-n(7)-)-methyltransferase, putative [Ricinus communis] |