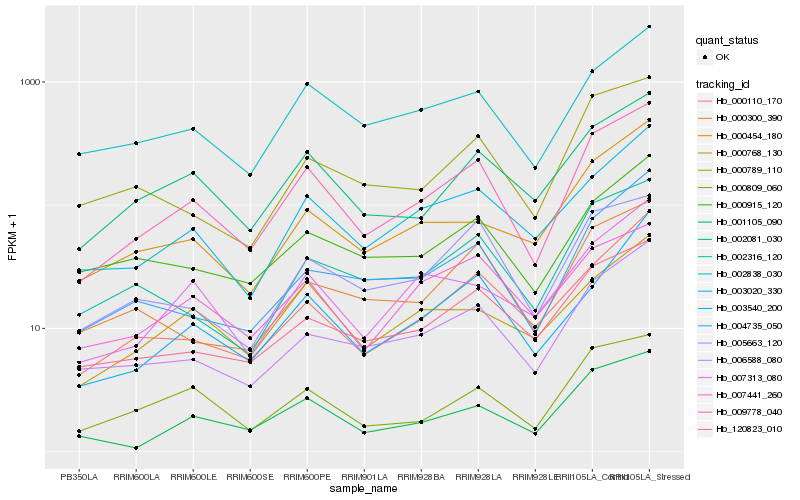
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009778_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105110611 [Populus euphratica] |
| 2 |
Hb_002081_030 |
0.1045474021 |
- |
- |
unnamed protein product [Coffea canephora] |
| 3 |
Hb_002838_030 |
0.1081519355 |
- |
- |
40S ribosomal protein S25A [Hevea brasiliensis] |
| 4 |
Hb_001105_090 |
0.1106673199 |
transcription factor |
TF Family: C3H |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_003020_330 |
0.1110182013 |
- |
- |
hypothetical protein B456_010G173100 [Gossypium raimondii] |
| 6 |
Hb_006588_080 |
0.1113140091 |
- |
- |
hypothetical protein JCGZ_25746 [Jatropha curcas] |
| 7 |
Hb_120823_010 |
0.1128264392 |
- |
- |
- |
| 8 |
Hb_000809_060 |
0.112837702 |
transcription factor |
TF Family: LOB |
LOB domain-containing 36 -like protein [Gossypium arboreum] |
| 9 |
Hb_000110_170 |
0.1196322511 |
- |
- |
PREDICTED: arginine biosynthesis bifunctional protein ArgJ, chloroplastic isoform X1 [Jatropha curcas] |
| 10 |
Hb_005663_120 |
0.1215271867 |
- |
- |
PREDICTED: uncharacterized protein LOC105635265 [Jatropha curcas] |
| 11 |
Hb_002316_120 |
0.1223635406 |
- |
- |
hypothetical protein POPTR_0015s08460g [Populus trichocarpa] |
| 12 |
Hb_007313_080 |
0.1228402025 |
- |
- |
hypothetical protein CICLE_v10002804mg [Citrus clementina] |
| 13 |
Hb_004735_050 |
0.1238239566 |
- |
- |
PREDICTED: uncharacterized protein LOC105639312 isoform X2 [Jatropha curcas] |
| 14 |
Hb_007441_260 |
0.124595144 |
- |
- |
PREDICTED: ATP synthase subunit delta', mitochondrial-like [Jatropha curcas] |
| 15 |
Hb_000789_110 |
0.1256223097 |
- |
- |
RecName: Full=Profilin-1; AltName: Full=Pollen allergen Hev b 8.0101; AltName: Allergen=Hev b 8.0101 [Hevea brasiliensis] |
| 16 |
Hb_000768_130 |
0.1275854277 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
PREDICTED: protein TRI1 [Jatropha curcas] |
| 17 |
Hb_000300_390 |
0.1281166509 |
- |
- |
BTB/POZ domain-containing protein KCTD9, putative [Ricinus communis] |
| 18 |
Hb_000915_120 |
0.1281264156 |
- |
- |
Peptidylprolyl cis/trans isomerase [Theobroma cacao] |
| 19 |
Hb_000454_180 |
0.1301520885 |
- |
- |
PREDICTED: uncharacterized protein LOC105134496 [Populus euphratica] |
| 20 |
Hb_003540_200 |
0.1304366121 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA3-like [Jatropha curcas] |