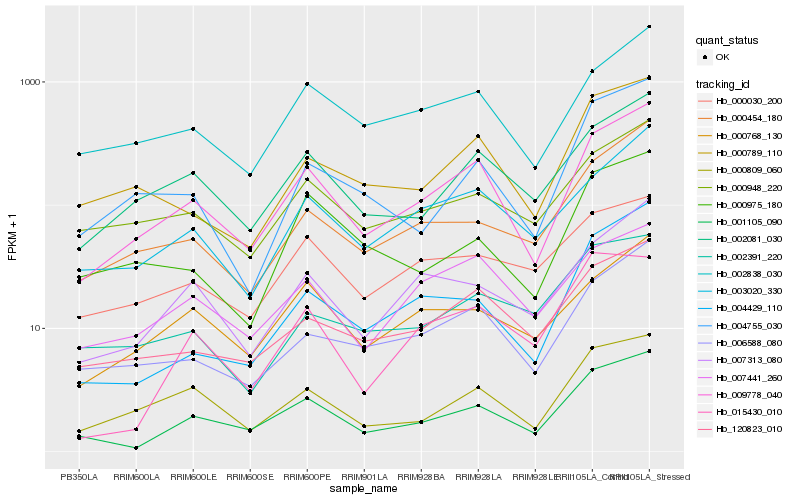
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001105_090 |
0.0 |
transcription factor |
TF Family: C3H |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_009778_040 |
0.1106673199 |
- |
- |
PREDICTED: uncharacterized protein LOC105110611 [Populus euphratica] |
| 3 |
Hb_004429_110 |
0.1395867255 |
- |
- |
PREDICTED: protein SPIRAL1-like 1 [Populus euphratica] |
| 4 |
Hb_007313_080 |
0.1400735232 |
- |
- |
hypothetical protein CICLE_v10002804mg [Citrus clementina] |
| 5 |
Hb_120823_010 |
0.1450195876 |
- |
- |
- |
| 6 |
Hb_000809_060 |
0.1460135514 |
transcription factor |
TF Family: LOB |
LOB domain-containing 36 -like protein [Gossypium arboreum] |
| 7 |
Hb_002081_030 |
0.1465680036 |
- |
- |
unnamed protein product [Coffea canephora] |
| 8 |
Hb_000454_180 |
0.1472897228 |
- |
- |
PREDICTED: uncharacterized protein LOC105134496 [Populus euphratica] |
| 9 |
Hb_015430_010 |
0.1475216877 |
- |
- |
PREDICTED: uncharacterized protein LOC105125506 isoform X2 [Populus euphratica] |
| 10 |
Hb_003020_330 |
0.1520059012 |
- |
- |
hypothetical protein B456_010G173100 [Gossypium raimondii] |
| 11 |
Hb_006588_080 |
0.1542391795 |
- |
- |
hypothetical protein JCGZ_25746 [Jatropha curcas] |
| 12 |
Hb_000768_130 |
0.1560998853 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
PREDICTED: protein TRI1 [Jatropha curcas] |
| 13 |
Hb_000948_220 |
0.1561162613 |
- |
- |
hypothetical protein CICLE_v10013189mg [Citrus clementina] |
| 14 |
Hb_002838_030 |
0.1590198253 |
- |
- |
40S ribosomal protein S25A [Hevea brasiliensis] |
| 15 |
Hb_000030_200 |
0.1607865086 |
- |
- |
PREDICTED: early nodulin-93-like [Populus euphratica] |
| 16 |
Hb_000789_110 |
0.1651312084 |
- |
- |
RecName: Full=Profilin-1; AltName: Full=Pollen allergen Hev b 8.0101; AltName: Allergen=Hev b 8.0101 [Hevea brasiliensis] |
| 17 |
Hb_000975_180 |
0.1656946124 |
- |
- |
Uncharacterized protein TCM_041879 [Theobroma cacao] |
| 18 |
Hb_002391_220 |
0.1682052272 |
- |
- |
PREDICTED: UPF0548 protein At2g17695 [Jatropha curcas] |
| 19 |
Hb_007441_260 |
0.1687217554 |
- |
- |
PREDICTED: ATP synthase subunit delta', mitochondrial-like [Jatropha curcas] |
| 20 |
Hb_004755_030 |
0.1706181413 |
- |
- |
EG964 [Manihot glaziovii] |