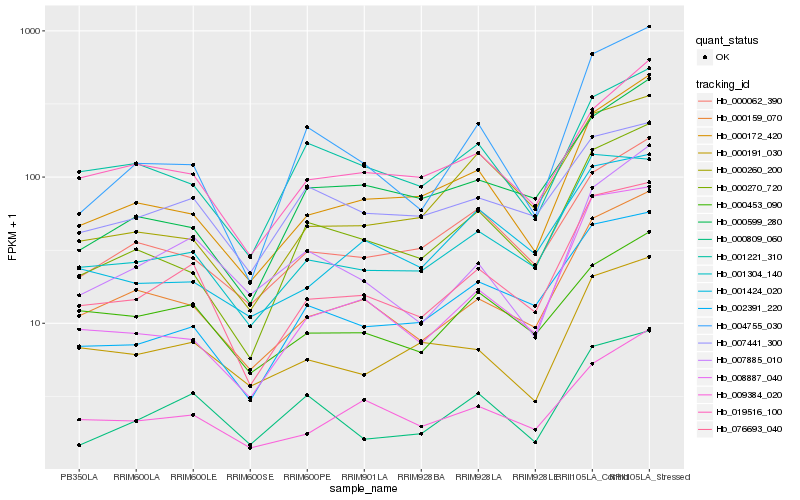
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_076693_040 |
0.0 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 5-B [Tarenaya hassleriana] |
| 2 |
Hb_001304_140 |
0.0767453846 |
- |
- |
PREDICTED: transmembrane protein 256 homolog [Jatropha curcas] |
| 3 |
Hb_000159_070 |
0.0877256431 |
- |
- |
JHL03K20.4 [Jatropha curcas] |
| 4 |
Hb_002391_220 |
0.1074250777 |
- |
- |
PREDICTED: UPF0548 protein At2g17695 [Jatropha curcas] |
| 5 |
Hb_009384_020 |
0.1109579724 |
- |
- |
hypothetical protein JCGZ_24168 [Jatropha curcas] |
| 6 |
Hb_000172_420 |
0.115920722 |
- |
- |
60S ribosomal protein L30, putative [Ricinus communis] |
| 7 |
Hb_000270_720 |
0.1164072874 |
- |
- |
hypothetical protein POPTR_0015s08460g [Populus trichocarpa] |
| 8 |
Hb_000062_390 |
0.1185353123 |
- |
- |
PREDICTED: protein Iojap-related, mitochondrial [Jatropha curcas] |
| 9 |
Hb_019516_100 |
0.1221883888 |
- |
- |
40S ribosomal protein S14, putative [Ricinus communis] |
| 10 |
Hb_000191_030 |
0.1228719447 |
- |
- |
hypothetical protein POPTR_0012s07830g [Populus trichocarpa] |
| 11 |
Hb_000809_060 |
0.1233817304 |
transcription factor |
TF Family: LOB |
LOB domain-containing 36 -like protein [Gossypium arboreum] |
| 12 |
Hb_007441_300 |
0.1269578418 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 13 |
Hb_008887_040 |
0.1269678086 |
- |
- |
Ribonuclease P family protein / Rpp14 family protein isoform 1 [Theobroma cacao] |
| 14 |
Hb_000260_200 |
0.1300769599 |
- |
- |
- |
| 15 |
Hb_004755_030 |
0.1315250355 |
- |
- |
EG964 [Manihot glaziovii] |
| 16 |
Hb_001221_310 |
0.1322476709 |
- |
- |
sec61 gamma subunit, putative [Ricinus communis] |
| 17 |
Hb_001424_020 |
0.1326618445 |
- |
- |
PREDICTED: AP-1 complex subunit sigma-2 isoform X2 [Solanum lycopersicum] |
| 18 |
Hb_000453_090 |
0.1328736593 |
- |
- |
mitochondrial import inner membrane translocase subunit Tim9 [Jatropha curcas] |
| 19 |
Hb_007885_010 |
0.1362358894 |
- |
- |
60S ribosomal protein L24-1 isoform 1 [Theobroma cacao] |
| 20 |
Hb_000599_280 |
0.1364417522 |
- |
- |
PREDICTED: 60S ribosomal protein L14-1 [Jatropha curcas] |