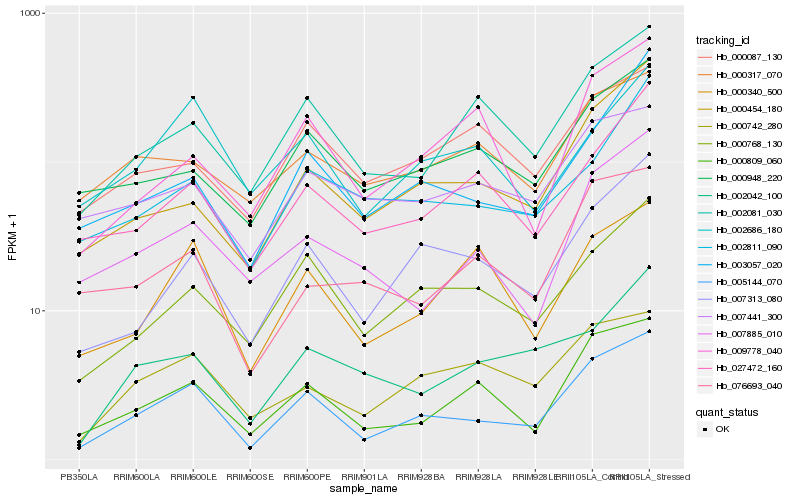
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005144_070 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000809_060 |
0.1232671126 |
transcription factor |
TF Family: LOB |
LOB domain-containing 36 -like protein [Gossypium arboreum] |
| 3 |
Hb_007313_080 |
0.1357851049 |
- |
- |
hypothetical protein CICLE_v10002804mg [Citrus clementina] |
| 4 |
Hb_000768_130 |
0.1451445552 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
PREDICTED: protein TRI1 [Jatropha curcas] |
| 5 |
Hb_002081_030 |
0.1508491655 |
- |
- |
unnamed protein product [Coffea canephora] |
| 6 |
Hb_000454_180 |
0.1530179652 |
- |
- |
PREDICTED: uncharacterized protein LOC105134496 [Populus euphratica] |
| 7 |
Hb_000742_280 |
0.1542389825 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 8 |
Hb_003057_020 |
0.1584624055 |
- |
- |
40S ribosomal protein S23, putative [Ricinus communis] |
| 9 |
Hb_000948_220 |
0.1609524849 |
- |
- |
hypothetical protein CICLE_v10013189mg [Citrus clementina] |
| 10 |
Hb_007885_010 |
0.1614298032 |
- |
- |
60S ribosomal protein L24-1 isoform 1 [Theobroma cacao] |
| 11 |
Hb_027472_160 |
0.1656421962 |
- |
- |
PREDICTED: proteasome subunit alpha type-2-A [Jatropha curcas] |
| 12 |
Hb_000340_500 |
0.1662701225 |
- |
- |
PREDICTED: aldo-keto reductase-like [Jatropha curcas] |
| 13 |
Hb_076693_040 |
0.1713472008 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 5-B [Tarenaya hassleriana] |
| 14 |
Hb_002042_100 |
0.1727419572 |
- |
- |
PREDICTED: uncharacterized protein LOC105647565 [Jatropha curcas] |
| 15 |
Hb_002811_090 |
0.1739898448 |
- |
- |
40S ribosomal protein S24A [Hevea brasiliensis] |
| 16 |
Hb_007441_300 |
0.1743929206 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 17 |
Hb_009778_040 |
0.175283272 |
- |
- |
PREDICTED: uncharacterized protein LOC105110611 [Populus euphratica] |
| 18 |
Hb_000317_070 |
0.1775072673 |
- |
- |
PREDICTED: uncharacterized protein LOC103942205 isoform X2 [Pyrus x bretschneideri] |
| 19 |
Hb_002686_180 |
0.1777617706 |
- |
- |
PREDICTED: early nodulin-93-like [Populus euphratica] |
| 20 |
Hb_000087_130 |
0.1781143499 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 6, mitochondrial [Jatropha curcas] |