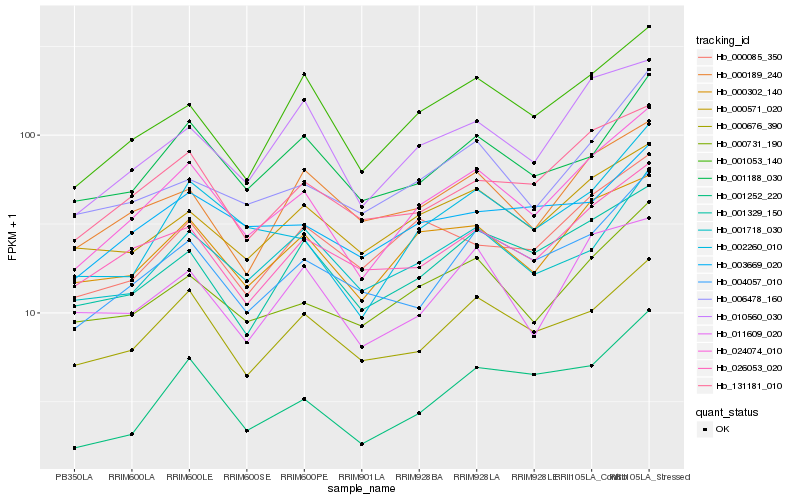
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_024074_010 |
0.0 |
- |
- |
PREDICTED: cytochrome c [Jatropha curcas] |
| 2 |
Hb_000302_140 |
0.0818849111 |
- |
- |
PREDICTED: probable NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 5, mitochondrial [Jatropha curcas] |
| 3 |
Hb_002260_010 |
0.0830390155 |
- |
- |
PREDICTED: coiled-coil-helix-coiled-coil-helix domain-containing protein 10, mitochondrial [Sesamum indicum] |
| 4 |
Hb_000731_190 |
0.085929777 |
- |
- |
PREDICTED: cardiolipin synthase (CMP-forming), mitochondrial [Jatropha curcas] |
| 5 |
Hb_001053_140 |
0.0903635484 |
- |
- |
PREDICTED: ATP synthase subunit O, mitochondrial [Jatropha curcas] |
| 6 |
Hb_004057_010 |
0.091801635 |
- |
- |
PREDICTED: uncharacterized protein LOC105649313 [Jatropha curcas] |
| 7 |
Hb_131181_010 |
0.0920072194 |
- |
- |
transcription initiation factor iia (tfiia), gamma chain, putative [Ricinus communis] |
| 8 |
Hb_026053_020 |
0.0927028915 |
- |
- |
PREDICTED: uncharacterized protein LOC105643718 [Jatropha curcas] |
| 9 |
Hb_010560_030 |
0.0938838458 |
- |
- |
PREDICTED: mitochondrial pyruvate carrier 4-like [Jatropha curcas] |
| 10 |
Hb_000676_390 |
0.0944477837 |
- |
- |
PREDICTED: protein FMP32, mitochondrial [Jatropha curcas] |
| 11 |
Hb_001329_150 |
0.0976805702 |
- |
- |
PREDICTED: autophagy-related protein 8i-like [Jatropha curcas] |
| 12 |
Hb_000571_020 |
0.0989712304 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 4 isoform X1 [Jatropha curcas] |
| 13 |
Hb_001188_030 |
0.099632182 |
- |
- |
PREDICTED: proteasome subunit beta type-4-like [Jatropha curcas] |
| 14 |
Hb_001718_030 |
0.1000655503 |
- |
- |
PREDICTED: calcineurin B-like protein 9 isoform X1 [Jatropha curcas] |
| 15 |
Hb_001252_220 |
0.1029859206 |
- |
- |
PREDICTED: uncharacterized protein LOC105640647 [Jatropha curcas] |
| 16 |
Hb_006478_160 |
0.1071663504 |
- |
- |
20S proteasome beta subunit D1 [Hevea brasiliensis] |
| 17 |
Hb_000085_350 |
0.1076037236 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM22-2-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_000189_240 |
0.1091348406 |
- |
- |
PREDICTED: PITH domain-containing protein At3g04780 [Jatropha curcas] |
| 19 |
Hb_011609_020 |
0.1098068015 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_003669_020 |
0.1100937859 |
- |
- |
PREDICTED: high mobility group nucleosome-binding domain-containing protein 5 [Populus euphratica] |