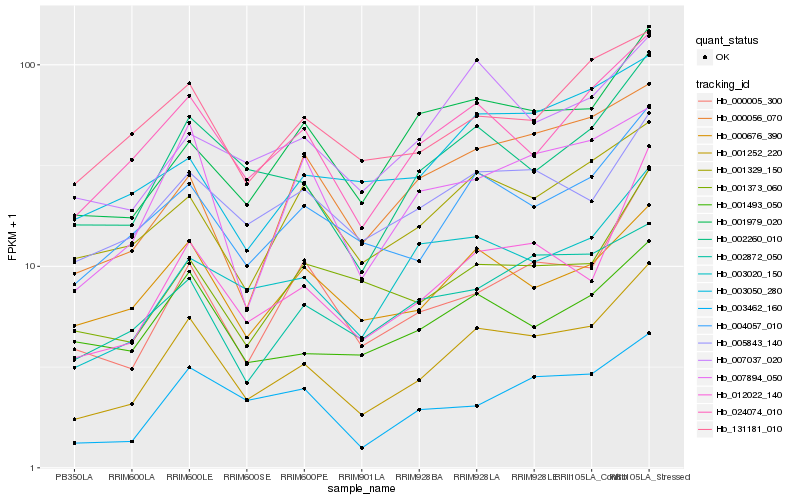
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001252_220 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105640647 [Jatropha curcas] |
| 2 |
Hb_002260_010 |
0.0994869244 |
- |
- |
PREDICTED: coiled-coil-helix-coiled-coil-helix domain-containing protein 10, mitochondrial [Sesamum indicum] |
| 3 |
Hb_012022_140 |
0.1016629538 |
- |
- |
ADP,ATP carrier protein, putative [Ricinus communis] |
| 4 |
Hb_004057_010 |
0.1024104918 |
- |
- |
PREDICTED: uncharacterized protein LOC105649313 [Jatropha curcas] |
| 5 |
Hb_024074_010 |
0.1029859206 |
- |
- |
PREDICTED: cytochrome c [Jatropha curcas] |
| 6 |
Hb_000005_300 |
0.1176204751 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_003020_150 |
0.1193563698 |
- |
- |
tropinone reductase, putative [Ricinus communis] |
| 8 |
Hb_003462_160 |
0.1240856788 |
transcription factor |
TF Family: GNAT |
predicted protein [Arabidopsis lyrata subsp. lyrata] |
| 9 |
Hb_001979_020 |
0.1251688614 |
- |
- |
hypothetical protein OsJ_31823 [Oryza sativa Japonica Group] |
| 10 |
Hb_001329_150 |
0.1256154118 |
- |
- |
PREDICTED: autophagy-related protein 8i-like [Jatropha curcas] |
| 11 |
Hb_000676_390 |
0.1274669935 |
- |
- |
PREDICTED: protein FMP32, mitochondrial [Jatropha curcas] |
| 12 |
Hb_007894_050 |
0.1288290004 |
- |
- |
PREDICTED: reactive Intermediate Deaminase A, chloroplastic [Jatropha curcas] |
| 13 |
Hb_002872_050 |
0.1309274421 |
- |
- |
PREDICTED: thioredoxin-like 3-2, chloroplastic [Jatropha curcas] |
| 14 |
Hb_005843_140 |
0.1316106885 |
- |
- |
Red chlorophyll catabolite reductase, chloroplast precursor, putative [Ricinus communis] |
| 15 |
Hb_001493_050 |
0.1326852871 |
- |
- |
PREDICTED: uncharacterized protein LOC105631312 [Jatropha curcas] |
| 16 |
Hb_001373_060 |
0.1329621907 |
- |
- |
PREDICTED: structure-specific endonuclease subunit slx1 [Jatropha curcas] |
| 17 |
Hb_131181_010 |
0.1330468905 |
- |
- |
transcription initiation factor iia (tfiia), gamma chain, putative [Ricinus communis] |
| 18 |
Hb_007037_020 |
0.1333186986 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 78 [Jatropha curcas] |
| 19 |
Hb_000056_070 |
0.1339230272 |
- |
- |
- |
| 20 |
Hb_003050_280 |
0.1359754576 |
transcription factor |
TF Family: HMG |
DNA-binding protein MNB1B, putative [Ricinus communis] |