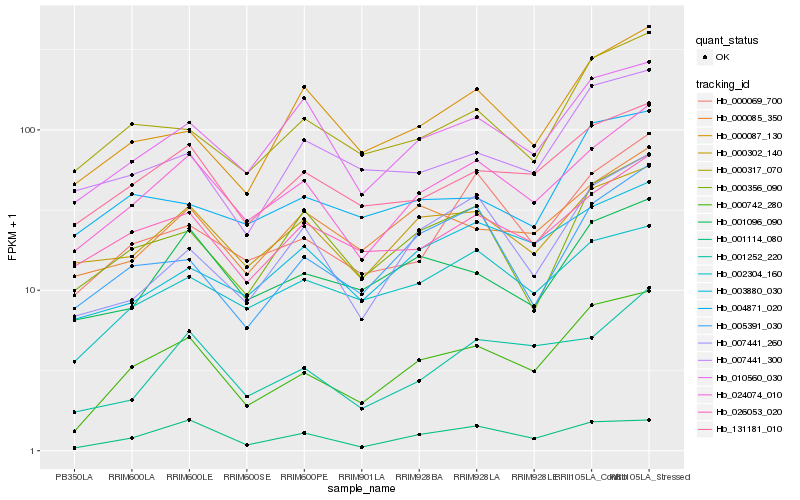
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000742_280 |
0.0 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 2 |
Hb_024074_010 |
0.1160148785 |
- |
- |
PREDICTED: cytochrome c [Jatropha curcas] |
| 3 |
Hb_131181_010 |
0.1275459765 |
- |
- |
transcription initiation factor iia (tfiia), gamma chain, putative [Ricinus communis] |
| 4 |
Hb_001114_080 |
0.1300858905 |
- |
- |
hypothetical protein PRUPE_ppa019021mg, partial [Prunus persica] |
| 5 |
Hb_000317_070 |
0.1320581226 |
- |
- |
PREDICTED: uncharacterized protein LOC103942205 isoform X2 [Pyrus x bretschneideri] |
| 6 |
Hb_010560_030 |
0.1380011753 |
- |
- |
PREDICTED: mitochondrial pyruvate carrier 4-like [Jatropha curcas] |
| 7 |
Hb_000069_700 |
0.1408036857 |
- |
- |
hypothetical protein B456_005G003200 [Gossypium raimondii] |
| 8 |
Hb_002304_160 |
0.1438261405 |
- |
- |
PREDICTED: nudix hydrolase 15, mitochondrial-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_001096_090 |
0.1447758792 |
transcription factor |
TF Family: Whirly |
PREDICTED: single-stranded DNA-bindig protein WHY2, mitochondrial [Jatropha curcas] |
| 10 |
Hb_000087_130 |
0.1448915325 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 6, mitochondrial [Jatropha curcas] |
| 11 |
Hb_005391_030 |
0.1453081886 |
- |
- |
putative cytidine/deoxycytidylate deaminase family protein [Zea mays] |
| 12 |
Hb_007441_300 |
0.1467478401 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 13 |
Hb_000085_350 |
0.1469629563 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM22-2-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_007441_260 |
0.1471288254 |
- |
- |
PREDICTED: ATP synthase subunit delta', mitochondrial-like [Jatropha curcas] |
| 15 |
Hb_004871_020 |
0.1480847894 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
PREDICTED: protein TRI1 [Jatropha curcas] |
| 16 |
Hb_001252_220 |
0.1491787298 |
- |
- |
PREDICTED: uncharacterized protein LOC105640647 [Jatropha curcas] |
| 17 |
Hb_003880_030 |
0.1494303146 |
- |
- |
PREDICTED: protein TIC 21, chloroplastic [Jatropha curcas] |
| 18 |
Hb_026053_020 |
0.149448118 |
- |
- |
PREDICTED: uncharacterized protein LOC105643718 [Jatropha curcas] |
| 19 |
Hb_000302_140 |
0.149518696 |
- |
- |
PREDICTED: probable NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 5, mitochondrial [Jatropha curcas] |
| 20 |
Hb_000356_090 |
0.1496000459 |
- |
- |
PREDICTED: uncharacterized protein At2g38710 [Jatropha curcas] |