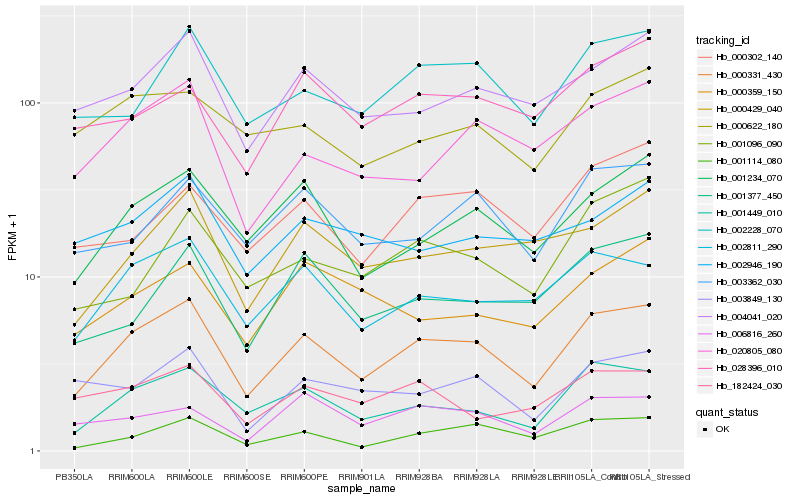
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000331_430 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105640242 [Jatropha curcas] |
| 2 |
Hb_001449_010 |
0.0989616788 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002811_290 |
0.1029207308 |
- |
- |
aldo/keto reductase, putative [Ricinus communis] |
| 4 |
Hb_001234_070 |
0.110659247 |
- |
- |
PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein Sgpp [Populus euphratica] |
| 5 |
Hb_002228_070 |
0.1154506726 |
- |
- |
inosine triphosphate pyrophosphatase, putative [Ricinus communis] |
| 6 |
Hb_004041_020 |
0.1172534261 |
- |
- |
ATP synthase D chain, mitochondrial, putative [Ricinus communis] |
| 7 |
Hb_000429_040 |
0.1210966684 |
- |
- |
PREDICTED: chaperone protein dnaJ 6 [Vitis vinifera] |
| 8 |
Hb_020805_080 |
0.1238558718 |
- |
- |
PREDICTED: uncharacterized protein LOC105635198 [Jatropha curcas] |
| 9 |
Hb_003362_030 |
0.1298865257 |
- |
- |
hypothetical protein POPTR_0017s08440g [Populus trichocarpa] |
| 10 |
Hb_001377_450 |
0.1302886939 |
- |
- |
PREDICTED: pyridoxine/pyridoxamine 5'-phosphate oxidase 2 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001114_080 |
0.1310039185 |
- |
- |
hypothetical protein PRUPE_ppa019021mg, partial [Prunus persica] |
| 12 |
Hb_000302_140 |
0.1337143007 |
- |
- |
PREDICTED: probable NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 5, mitochondrial [Jatropha curcas] |
| 13 |
Hb_006816_260 |
0.1374760397 |
- |
- |
hypothetical protein JCGZ_23244 [Jatropha curcas] |
| 14 |
Hb_003849_130 |
0.1393946008 |
- |
- |
PREDICTED: probable protein phosphatase 2C 14 [Jatropha curcas] |
| 15 |
Hb_182424_030 |
0.1397069639 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 8 [Vitis vinifera] |
| 16 |
Hb_028396_010 |
0.1410553107 |
- |
- |
PREDICTED: probable ATP synthase 24 kDa subunit, mitochondrial [Vitis vinifera] |
| 17 |
Hb_000622_180 |
0.1416490883 |
- |
- |
ubiquitin-conjugating enzyme E2, putative [Ricinus communis] |
| 18 |
Hb_001096_090 |
0.1417190154 |
transcription factor |
TF Family: Whirly |
PREDICTED: single-stranded DNA-bindig protein WHY2, mitochondrial [Jatropha curcas] |
| 19 |
Hb_000359_150 |
0.141853134 |
- |
- |
PREDICTED: protein Mpv17-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_002946_190 |
0.1433699142 |
- |
- |
PREDICTED: nudix hydrolase 26, chloroplastic [Jatropha curcas] |