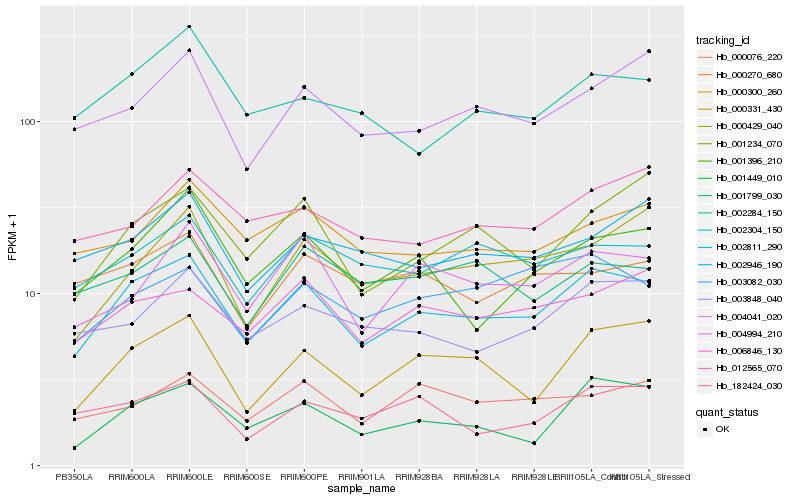
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002811_290 |
0.0 |
- |
- |
aldo/keto reductase, putative [Ricinus communis] |
| 2 |
Hb_000331_430 |
0.1029207308 |
- |
- |
PREDICTED: uncharacterized protein LOC105640242 [Jatropha curcas] |
| 3 |
Hb_004994_210 |
0.1078837747 |
- |
- |
phosphomannomutase family protein [Populus trichocarpa] |
| 4 |
Hb_006846_130 |
0.1096704006 |
- |
- |
PREDICTED: carbonyl reductase [NADPH] 1-like [Jatropha curcas] |
| 5 |
Hb_001396_210 |
0.1106247551 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_003848_040 |
0.1111968896 |
- |
- |
PREDICTED: phosphatidylinositol-glycan biosynthesis class X protein isoform X1 [Jatropha curcas] |
| 7 |
Hb_001449_010 |
0.1122529704 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_002304_150 |
0.1128008637 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_004041_020 |
0.1143790828 |
- |
- |
ATP synthase D chain, mitochondrial, putative [Ricinus communis] |
| 10 |
Hb_001234_070 |
0.1171058876 |
- |
- |
PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein Sgpp [Populus euphratica] |
| 11 |
Hb_000300_260 |
0.1184505619 |
- |
- |
PREDICTED: replication factor C subunit 3 [Jatropha curcas] |
| 12 |
Hb_000429_040 |
0.1195529103 |
- |
- |
PREDICTED: chaperone protein dnaJ 6 [Vitis vinifera] |
| 13 |
Hb_000076_220 |
0.1207353556 |
- |
- |
thioredoxin domain-containing protein, putative [Ricinus communis] |
| 14 |
Hb_003082_030 |
0.1216163877 |
- |
- |
PREDICTED: adenylate kinase, chloroplastic [Jatropha curcas] |
| 15 |
Hb_002284_150 |
0.1235220342 |
- |
- |
PREDICTED: uncharacterized protein LOC105635198 [Jatropha curcas] |
| 16 |
Hb_002946_190 |
0.1240588336 |
- |
- |
PREDICTED: nudix hydrolase 26, chloroplastic [Jatropha curcas] |
| 17 |
Hb_001799_030 |
0.1243841284 |
- |
- |
PREDICTED: ELMO domain-containing protein C-like [Jatropha curcas] |
| 18 |
Hb_000270_680 |
0.1243915171 |
- |
- |
hypothetical protein POPTR_0015s08420g [Populus trichocarpa] |
| 19 |
Hb_182424_030 |
0.1249889951 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 8 [Vitis vinifera] |
| 20 |
Hb_012565_070 |
0.1256305473 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 17B [Jatropha curcas] |