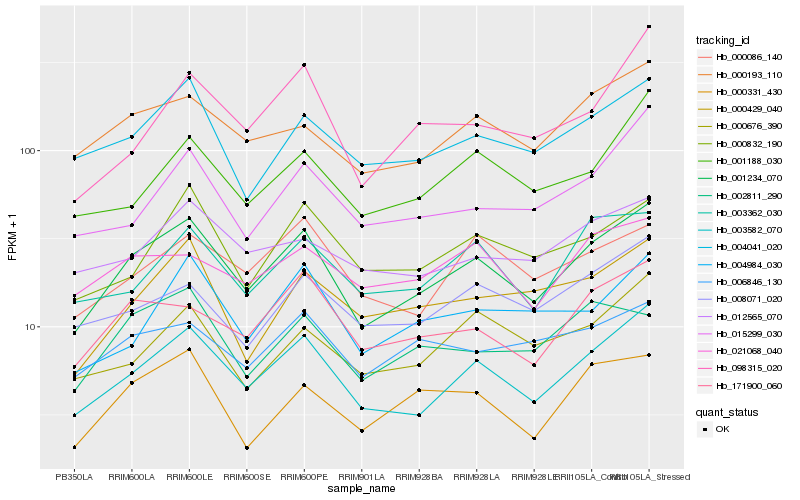
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001234_070 |
0.0 |
- |
- |
PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein Sgpp [Populus euphratica] |
| 2 |
Hb_003582_070 |
0.0558673973 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_171900_060 |
0.0975285655 |
- |
- |
PREDICTED: uncharacterized protein LOC105643368 [Jatropha curcas] |
| 4 |
Hb_000193_110 |
0.0977769839 |
- |
- |
PREDICTED: SKP1-like protein 1B [Jatropha curcas] |
| 5 |
Hb_004041_020 |
0.1021038932 |
- |
- |
ATP synthase D chain, mitochondrial, putative [Ricinus communis] |
| 6 |
Hb_003362_030 |
0.1067979397 |
- |
- |
hypothetical protein POPTR_0017s08440g [Populus trichocarpa] |
| 7 |
Hb_000832_190 |
0.1088265718 |
- |
- |
Rab3 [Hevea brasiliensis] |
| 8 |
Hb_000331_430 |
0.110659247 |
- |
- |
PREDICTED: uncharacterized protein LOC105640242 [Jatropha curcas] |
| 9 |
Hb_000086_140 |
0.1111283826 |
- |
- |
PREDICTED: probable arabinose 5-phosphate isomerase [Jatropha curcas] |
| 10 |
Hb_004984_030 |
0.1114714655 |
- |
- |
protein with unknown function [Ricinus communis] |
| 11 |
Hb_008071_020 |
0.1130709792 |
- |
- |
PREDICTED: serine/threonine-protein kinase AFC1 [Jatropha curcas] |
| 12 |
Hb_000429_040 |
0.1130970321 |
- |
- |
PREDICTED: chaperone protein dnaJ 6 [Vitis vinifera] |
| 13 |
Hb_001188_030 |
0.1131360789 |
- |
- |
PREDICTED: proteasome subunit beta type-4-like [Jatropha curcas] |
| 14 |
Hb_000676_390 |
0.1153714097 |
- |
- |
PREDICTED: protein FMP32, mitochondrial [Jatropha curcas] |
| 15 |
Hb_012565_070 |
0.1158797843 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 17B [Jatropha curcas] |
| 16 |
Hb_015299_030 |
0.1159202564 |
- |
- |
PREDICTED: proteasome subunit alpha type-7 [Jatropha curcas] |
| 17 |
Hb_006846_130 |
0.1159924232 |
- |
- |
PREDICTED: carbonyl reductase [NADPH] 1-like [Jatropha curcas] |
| 18 |
Hb_098315_020 |
0.1164190293 |
- |
- |
hypothetical protein M569_15712 [Genlisea aurea] |
| 19 |
Hb_002811_290 |
0.1171058876 |
- |
- |
aldo/keto reductase, putative [Ricinus communis] |
| 20 |
Hb_021068_040 |
0.118770619 |
- |
- |
PREDICTED: XIAP-associated factor 1 [Jatropha curcas] |