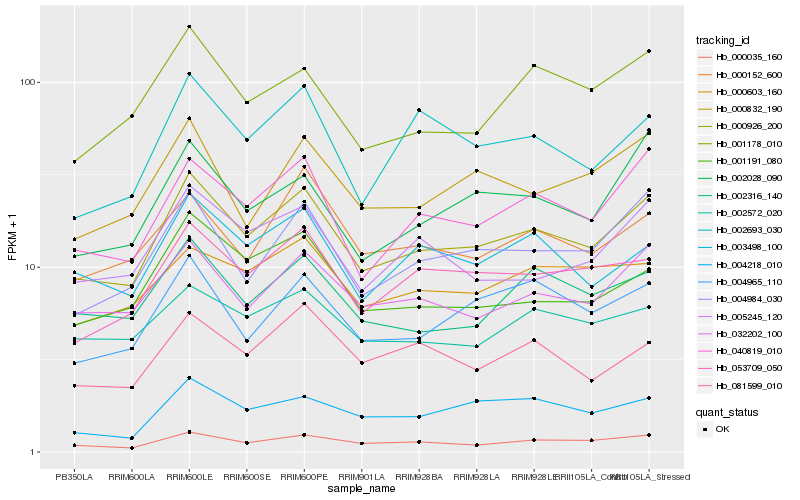
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000926_200 |
0.0 |
transcription factor |
TF Family: Rcd1-like |
PREDICTED: cell differentiation protein RCD1 homolog isoform X1 [Jatropha curcas] |
| 2 |
Hb_000035_160 |
0.0696206548 |
- |
- |
PREDICTED: formin-like protein 18 [Jatropha curcas] |
| 3 |
Hb_040819_010 |
0.0697516042 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 4 |
Hb_032202_100 |
0.0743432201 |
desease resistance |
Gene Name: ArsA_ATPase |
arsenical pump-driving atpase, putative [Ricinus communis] |
| 5 |
Hb_004984_030 |
0.0793793733 |
- |
- |
protein with unknown function [Ricinus communis] |
| 6 |
Hb_053709_050 |
0.0819970293 |
- |
- |
PREDICTED: nudix hydrolase 23, chloroplastic isoform X2 [Jatropha curcas] |
| 7 |
Hb_002316_140 |
0.0885732113 |
- |
- |
PREDICTED: rac-like GTP-binding protein 3 [Populus euphratica] |
| 8 |
Hb_003498_100 |
0.0905216889 |
- |
- |
component of oligomeric golgi complex, putative [Ricinus communis] |
| 9 |
Hb_005245_120 |
0.0915620992 |
- |
- |
XPA-binding protein, putative [Ricinus communis] |
| 10 |
Hb_000832_190 |
0.0924878934 |
- |
- |
Rab3 [Hevea brasiliensis] |
| 11 |
Hb_004965_110 |
0.0930474659 |
- |
- |
PREDICTED: folylpolyglutamate synthase [Jatropha curcas] |
| 12 |
Hb_001191_080 |
0.0940962415 |
- |
- |
protein with unknown function [Ricinus communis] |
| 13 |
Hb_001178_010 |
0.094471452 |
- |
- |
PREDICTED: uncharacterized protein LOC105629461 [Jatropha curcas] |
| 14 |
Hb_002572_020 |
0.0949667348 |
transcription factor |
TF Family: SBP |
hypothetical protein POPTR_0005s28010g [Populus trichocarpa] |
| 15 |
Hb_002028_090 |
0.0952165176 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase AIP2-like [Jatropha curcas] |
| 16 |
Hb_000603_160 |
0.0958148564 |
- |
- |
PREDICTED: fatty-acid-binding protein 2 [Jatropha curcas] |
| 17 |
Hb_002693_030 |
0.0969732904 |
- |
- |
PREDICTED: mitochondrial outer membrane protein porin of 34 kDa [Jatropha curcas] |
| 18 |
Hb_000152_600 |
0.0972140333 |
- |
- |
PREDICTED: protein jagunal homolog 1 [Jatropha curcas] |
| 19 |
Hb_081599_010 |
0.0976237813 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_004218_010 |
0.098340218 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |