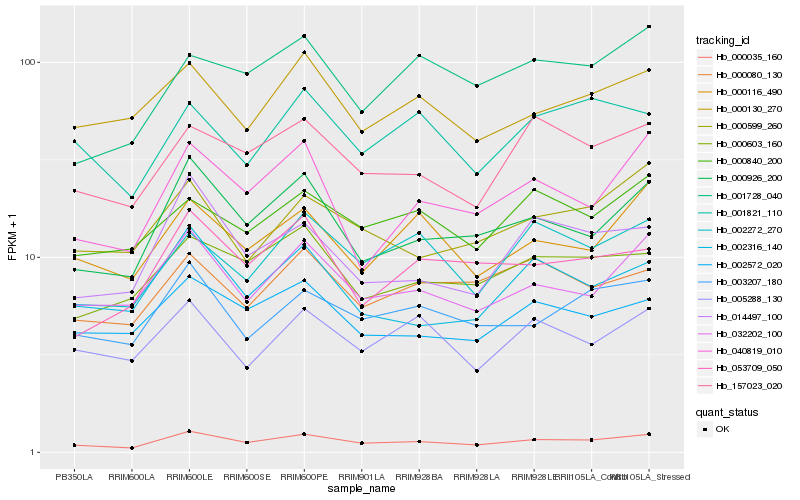
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000035_160 |
0.0 |
- |
- |
PREDICTED: formin-like protein 18 [Jatropha curcas] |
| 2 |
Hb_000926_200 |
0.0696206548 |
transcription factor |
TF Family: Rcd1-like |
PREDICTED: cell differentiation protein RCD1 homolog isoform X1 [Jatropha curcas] |
| 3 |
Hb_032202_100 |
0.0778797081 |
desease resistance |
Gene Name: ArsA_ATPase |
arsenical pump-driving atpase, putative [Ricinus communis] |
| 4 |
Hb_003207_180 |
0.0865965585 |
- |
- |
PREDICTED: uncharacterized protein LOC105645887 isoform X2 [Jatropha curcas] |
| 5 |
Hb_157023_020 |
0.0887652491 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 6 |
Hb_000116_490 |
0.0943276881 |
- |
- |
PREDICTED: WD repeat-containing protein 55 homolog [Jatropha curcas] |
| 7 |
Hb_000603_160 |
0.0950955149 |
- |
- |
PREDICTED: fatty-acid-binding protein 2 [Jatropha curcas] |
| 8 |
Hb_002572_020 |
0.0954104288 |
transcription factor |
TF Family: SBP |
hypothetical protein POPTR_0005s28010g [Populus trichocarpa] |
| 9 |
Hb_040819_010 |
0.0961603164 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 10 |
Hb_002272_270 |
0.0963919469 |
- |
- |
hypothetical protein POPTR_0013s02780g [Populus trichocarpa] |
| 11 |
Hb_002316_140 |
0.0967928119 |
- |
- |
PREDICTED: rac-like GTP-binding protein 3 [Populus euphratica] |
| 12 |
Hb_000130_270 |
0.0979722846 |
- |
- |
PREDICTED: malate dehydrogenase, mitochondrial [Jatropha curcas] |
| 13 |
Hb_005288_130 |
0.0987348605 |
- |
- |
protein with unknown function [Ricinus communis] |
| 14 |
Hb_014497_100 |
0.0990970572 |
- |
- |
PREDICTED: AT-rich interactive domain-containing protein 5 isoform X2 [Jatropha curcas] |
| 15 |
Hb_000080_130 |
0.0996794706 |
- |
- |
PREDICTED: TBC1 domain family member 22B [Jatropha curcas] |
| 16 |
Hb_000840_200 |
0.0998954946 |
- |
- |
PREDICTED: carbamoyl-phosphate synthase small chain, chloroplastic-like [Jatropha curcas] |
| 17 |
Hb_000599_260 |
0.1007135517 |
- |
- |
Vacuolar protein sorting-associated protein 2 like 3 [Glycine soja] |
| 18 |
Hb_053709_050 |
0.1031033892 |
- |
- |
PREDICTED: nudix hydrolase 23, chloroplastic isoform X2 [Jatropha curcas] |
| 19 |
Hb_001821_110 |
0.10412457 |
- |
- |
PREDICTED: ubiquinol oxidase, mitochondrial [Jatropha curcas] |
| 20 |
Hb_001728_040 |
0.1062128401 |
- |
- |
Aconitase/3-isopropylmalate dehydratase protein [Theobroma cacao] |