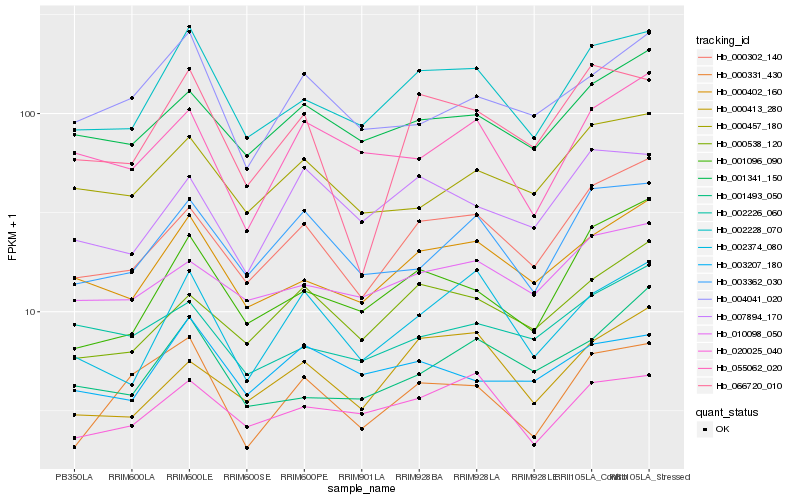
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002228_070 |
0.0 |
- |
- |
inosine triphosphate pyrophosphatase, putative [Ricinus communis] |
| 2 |
Hb_000402_160 |
0.0617446664 |
- |
- |
PREDICTED: ribose-phosphate pyrophosphokinase 4 isoform X1 [Jatropha curcas] |
| 3 |
Hb_020025_040 |
0.0885644738 |
- |
- |
sodium-bile acid cotransporter, putative [Ricinus communis] |
| 4 |
Hb_000302_140 |
0.0911223108 |
- |
- |
PREDICTED: probable NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 5, mitochondrial [Jatropha curcas] |
| 5 |
Hb_001096_090 |
0.0946852445 |
transcription factor |
TF Family: Whirly |
PREDICTED: single-stranded DNA-bindig protein WHY2, mitochondrial [Jatropha curcas] |
| 6 |
Hb_001341_150 |
0.0986880643 |
- |
- |
PREDICTED: proteasome subunit beta type-4-like [Jatropha curcas] |
| 7 |
Hb_010098_050 |
0.1007553403 |
- |
- |
PREDICTED: putative deoxyribonuclease tatdn3-A [Jatropha curcas] |
| 8 |
Hb_003362_030 |
0.1025982994 |
- |
- |
hypothetical protein POPTR_0017s08440g [Populus trichocarpa] |
| 9 |
Hb_002374_080 |
0.1052508971 |
- |
- |
PREDICTED: chloroplast processing peptidase-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_001493_050 |
0.107071619 |
- |
- |
PREDICTED: uncharacterized protein LOC105631312 [Jatropha curcas] |
| 11 |
Hb_003207_180 |
0.1074139514 |
- |
- |
PREDICTED: uncharacterized protein LOC105645887 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000457_180 |
0.1114303561 |
- |
- |
PREDICTED: cytochrome c1-2, heme protein, mitochondrial [Jatropha curcas] |
| 13 |
Hb_000538_120 |
0.1137066171 |
transcription factor |
TF Family: SET |
PREDICTED: protein SET DOMAIN GROUP 40 [Jatropha curcas] |
| 14 |
Hb_000331_430 |
0.1154506726 |
- |
- |
PREDICTED: uncharacterized protein LOC105640242 [Jatropha curcas] |
| 15 |
Hb_004041_020 |
0.1163199136 |
- |
- |
ATP synthase D chain, mitochondrial, putative [Ricinus communis] |
| 16 |
Hb_055062_020 |
0.1166939312 |
- |
- |
60S ribosomal L35a-3 -like protein [Gossypium arboreum] |
| 17 |
Hb_007894_170 |
0.1169339604 |
- |
- |
PREDICTED: UPF0136 membrane protein At2g26240-like [Jatropha curcas] |
| 18 |
Hb_002226_060 |
0.1169703455 |
- |
- |
PREDICTED: uncharacterized protein LOC105641377 [Jatropha curcas] |
| 19 |
Hb_066720_010 |
0.1174081919 |
- |
- |
PREDICTED: histone H2B [Jatropha curcas] |
| 20 |
Hb_000413_280 |
0.120591273 |
- |
- |
PREDICTED: probable UDP-3-O-acylglucosamine N-acyltransferase 2, mitochondrial [Jatropha curcas] |