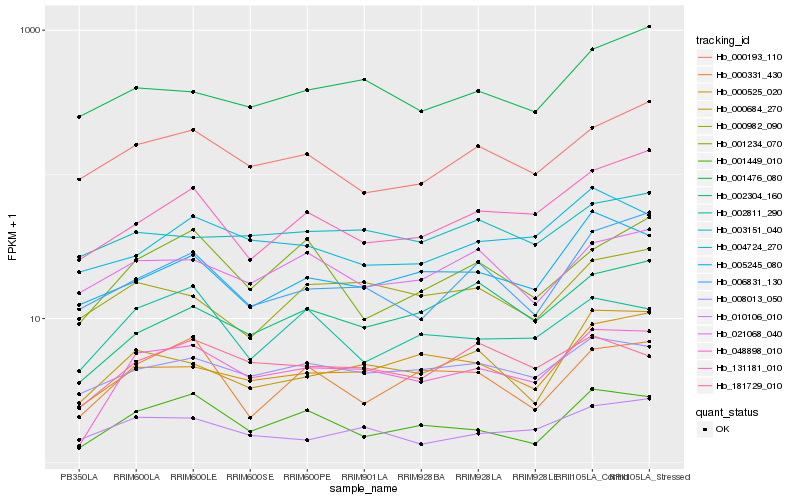
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_048898_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105116251 isoform X2 [Populus euphratica] |
| 2 |
Hb_001449_010 |
0.1293288719 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_008013_050 |
0.1348477743 |
transcription factor |
TF Family: PHD |
PREDICTED: PHD finger protein At1g33420 [Jatropha curcas] |
| 4 |
Hb_181729_010 |
0.1365421763 |
- |
- |
PREDICTED: DNA polymerase epsilon subunit 2 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000525_020 |
0.1380615691 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 6 |
Hb_002304_160 |
0.1413776644 |
- |
- |
PREDICTED: nudix hydrolase 15, mitochondrial-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_000684_270 |
0.1414796641 |
- |
- |
PREDICTED: probable DNA helicase MCM8 isoform X1 [Jatropha curcas] |
| 8 |
Hb_010106_010 |
0.1421847944 |
- |
- |
PREDICTED: cucumisin-like [Populus euphratica] |
| 9 |
Hb_005245_080 |
0.1462922343 |
- |
- |
PREDICTED: uncharacterized protein LOC105643645 isoform X2 [Jatropha curcas] |
| 10 |
Hb_131181_010 |
0.1464301704 |
- |
- |
transcription initiation factor iia (tfiia), gamma chain, putative [Ricinus communis] |
| 11 |
Hb_000193_110 |
0.1508258388 |
- |
- |
PREDICTED: SKP1-like protein 1B [Jatropha curcas] |
| 12 |
Hb_002811_290 |
0.150838903 |
- |
- |
aldo/keto reductase, putative [Ricinus communis] |
| 13 |
Hb_001476_080 |
0.1522533944 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 5A-like [Jatropha curcas] |
| 14 |
Hb_006831_130 |
0.1527454121 |
- |
- |
catalytic, putative [Ricinus communis] |
| 15 |
Hb_000331_430 |
0.154140467 |
- |
- |
PREDICTED: uncharacterized protein LOC105640242 [Jatropha curcas] |
| 16 |
Hb_004724_270 |
0.1543170846 |
- |
- |
hypothetical protein JCGZ_12706 [Jatropha curcas] |
| 17 |
Hb_021068_040 |
0.1558178674 |
- |
- |
PREDICTED: XIAP-associated factor 1 [Jatropha curcas] |
| 18 |
Hb_001234_070 |
0.1562218759 |
- |
- |
PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein Sgpp [Populus euphratica] |
| 19 |
Hb_003151_040 |
0.1568906299 |
transcription factor |
TF Family: Alfin-like |
zinc finger family protein [Populus trichocarpa] |
| 20 |
Hb_000982_090 |
0.1570494009 |
- |
- |
PREDICTED: zinc finger HIT domain-containing protein 3 [Jatropha curcas] |