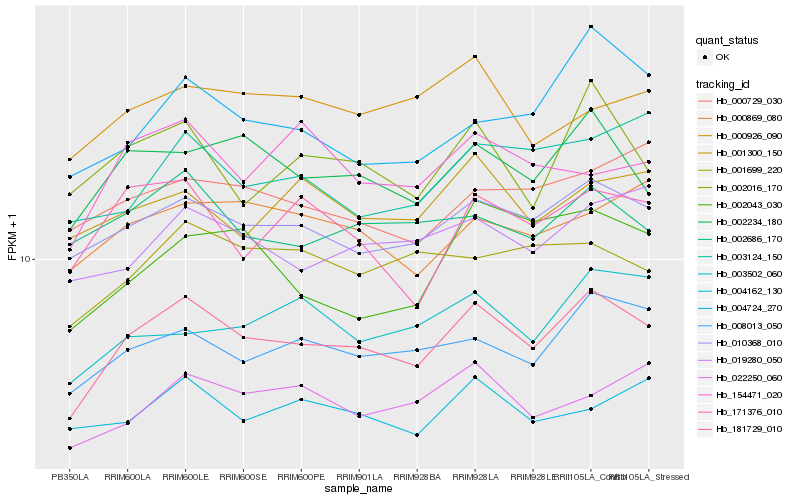
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_181729_010 |
0.0 |
- |
- |
PREDICTED: DNA polymerase epsilon subunit 2 isoform X1 [Jatropha curcas] |
| 2 |
Hb_010368_010 |
0.0803958723 |
- |
- |
RNA splicing protein mrs2, mitochondrial, putative [Ricinus communis] |
| 3 |
Hb_002234_180 |
0.0885188016 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_008013_050 |
0.0910583497 |
transcription factor |
TF Family: PHD |
PREDICTED: PHD finger protein At1g33420 [Jatropha curcas] |
| 5 |
Hb_002016_170 |
0.1023324075 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 6 |
Hb_019280_050 |
0.1060128502 |
- |
- |
PREDICTED: protein SAWADEE HOMEODOMAIN HOMOLOG 2 [Jatropha curcas] |
| 7 |
Hb_171376_010 |
0.1063470618 |
- |
- |
PREDICTED: uncharacterized protein LOC105630877 isoform X1 [Jatropha curcas] |
| 8 |
Hb_002686_170 |
0.1067846117 |
- |
- |
PREDICTED: uncharacterized protein LOC105632645 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001699_220 |
0.1089588925 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH121-like [Jatropha curcas] |
| 10 |
Hb_022250_060 |
0.111493146 |
- |
- |
PREDICTED: calcium-binding mitochondrial carrier protein SCaMC-1-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_004724_270 |
0.1116614639 |
- |
- |
hypothetical protein JCGZ_12706 [Jatropha curcas] |
| 12 |
Hb_000869_080 |
0.1121165202 |
- |
- |
PREDICTED: uncharacterized protein LOC105641084 isoform X2 [Jatropha curcas] |
| 13 |
Hb_001300_150 |
0.1138086362 |
transcription factor |
TF Family: TUB |
PREDICTED: tubby-like F-box protein 3 [Jatropha curcas] |
| 14 |
Hb_003124_150 |
0.1141306836 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 28-like [Jatropha curcas] |
| 15 |
Hb_003502_060 |
0.1142654072 |
- |
- |
PREDICTED: alpha-1,3/1,6-mannosyltransferase ALG2 [Jatropha curcas] |
| 16 |
Hb_004162_130 |
0.1160889192 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_002043_030 |
0.1162484477 |
- |
- |
hypothetical protein RCOM_1486170 [Ricinus communis] |
| 18 |
Hb_154471_020 |
0.1163323487 |
- |
- |
squalene synthetase [Euphorbia tirucalli] |
| 19 |
Hb_000729_030 |
0.1166270032 |
- |
- |
PREDICTED: dentin sialophosphoprotein [Jatropha curcas] |
| 20 |
Hb_000926_090 |
0.1166688038 |
- |
- |
PREDICTED: dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex 2, mitochondrial-like [Jatropha curcas] |