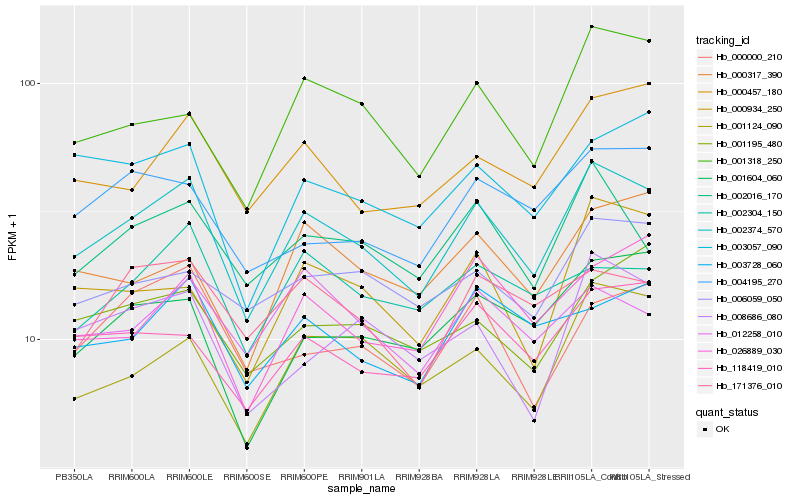
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002374_570 |
0.0 |
- |
- |
PREDICTED: vesicle-associated protein 1-2-like [Jatropha curcas] |
| 2 |
Hb_002016_170 |
0.0883549825 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 3 |
Hb_118419_010 |
0.0917320903 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 5 [Jatropha curcas] |
| 4 |
Hb_001124_090 |
0.0940415809 |
- |
- |
acyl carrier protein, putative [Ricinus communis] |
| 5 |
Hb_000457_180 |
0.0955299092 |
- |
- |
PREDICTED: cytochrome c1-2, heme protein, mitochondrial [Jatropha curcas] |
| 6 |
Hb_001318_250 |
0.0959248482 |
- |
- |
PREDICTED: uncharacterized protein LOC105642666 [Jatropha curcas] |
| 7 |
Hb_003728_060 |
0.0959906539 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 4-like [Jatropha curcas] |
| 8 |
Hb_003057_090 |
0.0970303933 |
- |
- |
ATP synthase D chain, mitochondrial, putative [Ricinus communis] |
| 9 |
Hb_171376_010 |
0.0973060547 |
- |
- |
PREDICTED: uncharacterized protein LOC105630877 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001604_060 |
0.0988642462 |
- |
- |
PREDICTED: TPR repeat-containing thioredoxin TTL1 [Jatropha curcas] |
| 11 |
Hb_026889_030 |
0.1005382022 |
- |
- |
protein with unknown function [Ricinus communis] |
| 12 |
Hb_006059_050 |
0.1006757598 |
- |
- |
PREDICTED: polyadenylate-binding protein 1 [Jatropha curcas] |
| 13 |
Hb_004195_270 |
0.1014706022 |
- |
- |
Ubiquitin-conjugating enzyme 16 [Theobroma cacao] |
| 14 |
Hb_002304_150 |
0.1016871496 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_008686_080 |
0.1018562764 |
- |
- |
PREDICTED: DNA-directed RNA polymerase V subunit 5A-like [Jatropha curcas] |
| 16 |
Hb_012258_010 |
0.1019831667 |
- |
- |
PREDICTED: cyclase-associated protein 1 [Jatropha curcas] |
| 17 |
Hb_000317_390 |
0.1023959444 |
- |
- |
PREDICTED: vesicle transport protein SFT2B [Jatropha curcas] |
| 18 |
Hb_001195_480 |
0.1024265753 |
- |
- |
cop9 complex subunit, putative [Ricinus communis] |
| 19 |
Hb_000000_210 |
0.1028852358 |
- |
- |
PREDICTED: F-box/FBD/LRR-repeat protein At1g13570-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_000934_250 |
0.1033770345 |
- |
- |
PREDICTED: probable arabinosyltransferase ARAD1 [Jatropha curcas] |