| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
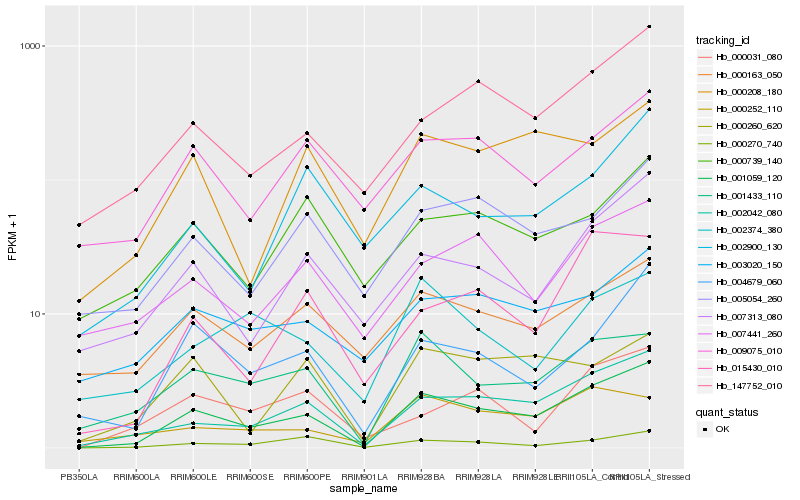
Hb_001059_120 |
0.0 |
- |
- |
NADH dehydrogenase, putative [Ricinus communis] |
| 2 |
Hb_002042_080 |
0.1196205503 |
rubber biosynthesis |
Gene Name: Mevalonate kinase |
mevalonate kinase [Hevea brasiliensis] |
| 3 |
Hb_001433_110 |
0.1654142368 |
- |
- |
PREDICTED: uncharacterized protein LOC105647469 isoform X1 [Jatropha curcas] |
| 4 |
Hb_147752_010 |
0.1716764699 |
- |
- |
PREDICTED: 40S ribosomal protein S9-2-like isoform X1 [Populus euphratica] |
| 5 |
Hb_004679_060 |
0.1744138015 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_007313_080 |
0.176527841 |
- |
- |
hypothetical protein CICLE_v10002804mg [Citrus clementina] |
| 7 |
Hb_000270_740 |
0.177681902 |
- |
- |
PREDICTED: probable inactive receptor kinase At2g26730 [Jatropha curcas] |
| 8 |
Hb_015430_010 |
0.1870207024 |
- |
- |
PREDICTED: uncharacterized protein LOC105125506 isoform X2 [Populus euphratica] |
| 9 |
Hb_003020_150 |
0.1885327981 |
- |
- |
tropinone reductase, putative [Ricinus communis] |
| 10 |
Hb_009075_010 |
0.1898250736 |
- |
- |
60S ribosomal protein L10B [Hevea brasiliensis] |
| 11 |
Hb_005054_260 |
0.1909943122 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 7 homolog A [Jatropha curcas] |
| 12 |
Hb_000163_050 |
0.1915049599 |
- |
- |
SPLICEOSOMAL protein U1A [Populus trichocarpa] |
| 13 |
Hb_007441_260 |
0.1935583232 |
- |
- |
PREDICTED: ATP synthase subunit delta', mitochondrial-like [Jatropha curcas] |
| 14 |
Hb_000208_180 |
0.197582435 |
- |
- |
PREDICTED: histone H2B-like [Populus euphratica] |
| 15 |
Hb_000252_110 |
0.2013331237 |
- |
- |
- |
| 16 |
Hb_002374_380 |
0.2042571487 |
- |
- |
Acyl-CoA N-acyltransferases (NAT) superfamily protein isoform 1 [Theobroma cacao] |
| 17 |
Hb_000031_080 |
0.2043786392 |
- |
- |
PREDICTED: uncharacterized protein LOC105637482 [Jatropha curcas] |
| 18 |
Hb_000739_140 |
0.2044548874 |
- |
- |
PREDICTED: proteasome subunit beta type-1 [Jatropha curcas] |
| 19 |
Hb_002900_130 |
0.2048758942 |
- |
- |
60S ribosomal L35a-3 -like protein [Gossypium arboreum] |
| 20 |
Hb_000260_620 |
0.2049839172 |
- |
- |
Thermosensitive gluconokinase, putative [Ricinus communis] |