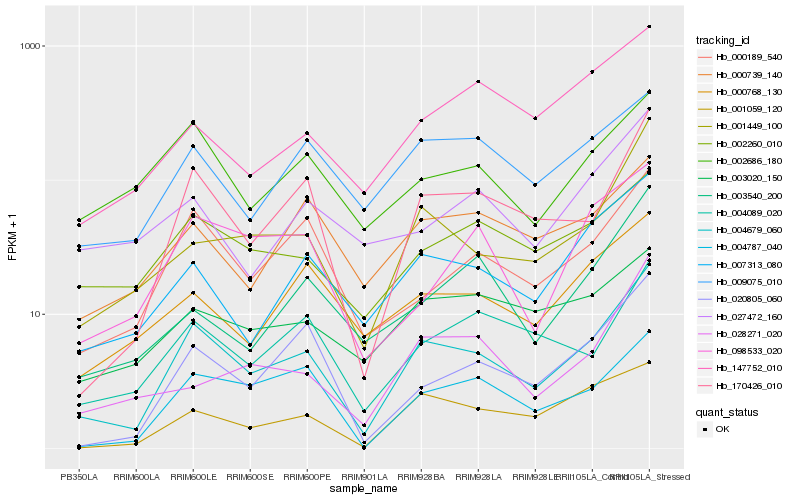
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004679_060 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_170426_010 |
0.1256804833 |
- |
- |
cytochrome b5 isoform Cb5-B [Vernicia fordii] |
| 3 |
Hb_001449_100 |
0.1561837816 |
- |
- |
PREDICTED: transcription factor bHLH149 [Jatropha curcas] |
| 4 |
Hb_007313_080 |
0.1596492543 |
- |
- |
hypothetical protein CICLE_v10002804mg [Citrus clementina] |
| 5 |
Hb_000189_540 |
0.1651885827 |
- |
- |
PREDICTED: tRNA(adenine(34)) deaminase, chloroplastic [Jatropha curcas] |
| 6 |
Hb_020805_060 |
0.1677877875 |
- |
- |
PREDICTED: protein S-acyltransferase 21 [Jatropha curcas] |
| 7 |
Hb_004787_040 |
0.1698430448 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001059_120 |
0.1744138015 |
- |
- |
NADH dehydrogenase, putative [Ricinus communis] |
| 9 |
Hb_004089_020 |
0.1813895574 |
- |
- |
PREDICTED: 3-phosphoinositide-dependent protein kinase 2 [Jatropha curcas] |
| 10 |
Hb_000768_130 |
0.1865521233 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
PREDICTED: protein TRI1 [Jatropha curcas] |
| 11 |
Hb_009075_010 |
0.1871491017 |
- |
- |
60S ribosomal protein L10B [Hevea brasiliensis] |
| 12 |
Hb_098533_020 |
0.1896363671 |
- |
- |
PREDICTED: zinc finger AN1 domain-containing stress-associated protein 12 [Jatropha curcas] |
| 13 |
Hb_002260_010 |
0.1932524029 |
- |
- |
PREDICTED: coiled-coil-helix-coiled-coil-helix domain-containing protein 10, mitochondrial [Sesamum indicum] |
| 14 |
Hb_028271_020 |
0.1939824057 |
- |
- |
acyl-CoA oxidase, putative [Ricinus communis] |
| 15 |
Hb_003540_200 |
0.1944065318 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA3-like [Jatropha curcas] |
| 16 |
Hb_002686_180 |
0.1944161031 |
- |
- |
PREDICTED: early nodulin-93-like [Populus euphratica] |
| 17 |
Hb_000739_140 |
0.1960036919 |
- |
- |
PREDICTED: proteasome subunit beta type-1 [Jatropha curcas] |
| 18 |
Hb_147752_010 |
0.1975784718 |
- |
- |
PREDICTED: 40S ribosomal protein S9-2-like isoform X1 [Populus euphratica] |
| 19 |
Hb_027472_160 |
0.1986596052 |
- |
- |
PREDICTED: proteasome subunit alpha type-2-A [Jatropha curcas] |
| 20 |
Hb_003020_150 |
0.2009488351 |
- |
- |
tropinone reductase, putative [Ricinus communis] |