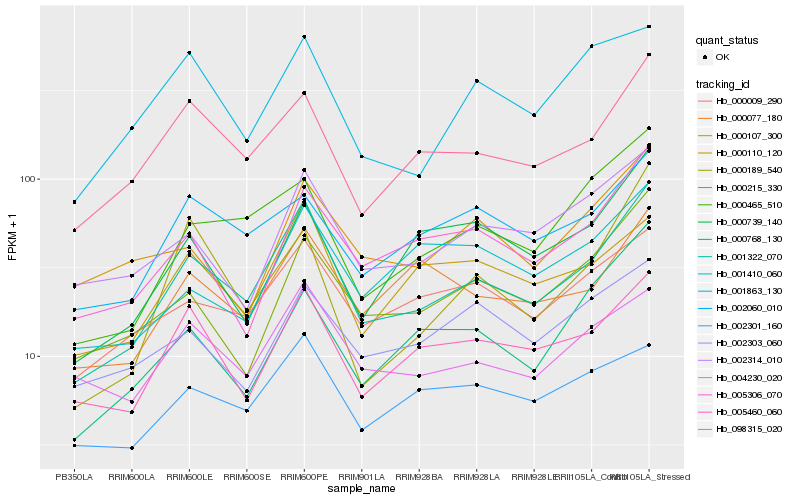
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001322_070 |
0.0 |
- |
- |
protein with unknown function [Ricinus communis] |
| 2 |
Hb_098315_020 |
0.1039389864 |
- |
- |
hypothetical protein M569_15712 [Genlisea aurea] |
| 3 |
Hb_005460_060 |
0.113015877 |
- |
- |
PREDICTED: proteasome assembly chaperone 2 [Jatropha curcas] |
| 4 |
Hb_000009_290 |
0.1189367266 |
- |
- |
PREDICTED: cold-regulated 413 plasma membrane protein 4-like [Jatropha curcas] |
| 5 |
Hb_000465_510 |
0.1191940643 |
- |
- |
PREDICTED: GTP-binding protein YPTM2 [Populus euphratica] |
| 6 |
Hb_000215_330 |
0.1194771986 |
- |
- |
PREDICTED: AP-4 complex subunit sigma [Jatropha curcas] |
| 7 |
Hb_005306_070 |
0.1197548609 |
- |
- |
PREDICTED: coatomer subunit zeta-1-like [Jatropha curcas] |
| 8 |
Hb_002314_010 |
0.1219756431 |
- |
- |
hypothetical protein JCGZ_15712 [Jatropha curcas] |
| 9 |
Hb_000077_180 |
0.1235064898 |
- |
- |
- |
| 10 |
Hb_004230_020 |
0.1241732128 |
- |
- |
PREDICTED: uncharacterized protein LOC105648105 [Jatropha curcas] |
| 11 |
Hb_000110_120 |
0.1249587587 |
- |
- |
PREDICTED: ras-related protein RABE1a [Fragaria vesca subsp. vesca] |
| 12 |
Hb_002060_010 |
0.1310055971 |
- |
- |
PREDICTED: uncharacterized protein LOC105642435 isoform X1 [Jatropha curcas] |
| 13 |
Hb_002301_160 |
0.1342251583 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g04780-like [Jatropha curcas] |
| 14 |
Hb_000107_300 |
0.1361460671 |
- |
- |
PREDICTED: AP-1 complex subunit sigma-2 [Jatropha curcas] |
| 15 |
Hb_000739_140 |
0.136965987 |
- |
- |
PREDICTED: proteasome subunit beta type-1 [Jatropha curcas] |
| 16 |
Hb_001863_130 |
0.1380409849 |
- |
- |
hypothetical protein ZEAMMB73_433257, partial [Zea mays] |
| 17 |
Hb_001410_060 |
0.1394521224 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 8 homolog A-like [Jatropha curcas] |
| 18 |
Hb_000768_130 |
0.1398432146 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
PREDICTED: protein TRI1 [Jatropha curcas] |
| 19 |
Hb_000189_540 |
0.1427696909 |
- |
- |
PREDICTED: tRNA(adenine(34)) deaminase, chloroplastic [Jatropha curcas] |
| 20 |
Hb_002303_060 |
0.145161528 |
- |
- |
conserved hypothetical protein [Ricinus communis] |