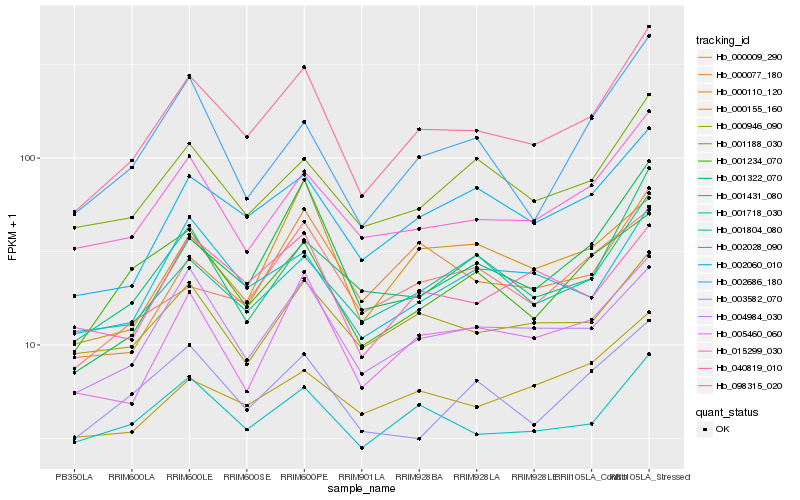
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_098315_020 |
0.0 |
- |
- |
hypothetical protein M569_15712 [Genlisea aurea] |
| 2 |
Hb_002060_010 |
0.0883480923 |
- |
- |
PREDICTED: uncharacterized protein LOC105642435 isoform X1 [Jatropha curcas] |
| 3 |
Hb_015299_030 |
0.0913648126 |
- |
- |
PREDICTED: proteasome subunit alpha type-7 [Jatropha curcas] |
| 4 |
Hb_005460_060 |
0.0949291445 |
- |
- |
PREDICTED: proteasome assembly chaperone 2 [Jatropha curcas] |
| 5 |
Hb_004984_030 |
0.0974146827 |
- |
- |
protein with unknown function [Ricinus communis] |
| 6 |
Hb_001188_030 |
0.1015942167 |
- |
- |
PREDICTED: proteasome subunit beta type-4-like [Jatropha curcas] |
| 7 |
Hb_001718_030 |
0.1037866203 |
- |
- |
PREDICTED: calcineurin B-like protein 9 isoform X1 [Jatropha curcas] |
| 8 |
Hb_001322_070 |
0.1039389864 |
- |
- |
protein with unknown function [Ricinus communis] |
| 9 |
Hb_001431_080 |
0.1048228004 |
- |
- |
40S ribosomal protein S11, putative [Ricinus communis] |
| 10 |
Hb_001804_080 |
0.1056975893 |
- |
- |
PREDICTED: aldo-keto reductase family 4 member C9-like [Jatropha curcas] |
| 11 |
Hb_002028_090 |
0.1088102481 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase AIP2-like [Jatropha curcas] |
| 12 |
Hb_003582_070 |
0.1108260912 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000946_090 |
0.1112514445 |
- |
- |
PREDICTED: cyclin-C1-2-like isoform X2 [Jatropha curcas] |
| 14 |
Hb_000077_180 |
0.1134589361 |
- |
- |
- |
| 15 |
Hb_001234_070 |
0.1164190293 |
- |
- |
PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein Sgpp [Populus euphratica] |
| 16 |
Hb_040819_010 |
0.1164707615 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 17 |
Hb_000155_160 |
0.1195670786 |
- |
- |
hypothetical protein POPTR_0011s05880g [Populus trichocarpa] |
| 18 |
Hb_002686_180 |
0.1206674504 |
- |
- |
PREDICTED: early nodulin-93-like [Populus euphratica] |
| 19 |
Hb_000009_290 |
0.1227386222 |
- |
- |
PREDICTED: cold-regulated 413 plasma membrane protein 4-like [Jatropha curcas] |
| 20 |
Hb_000110_120 |
0.127640414 |
- |
- |
PREDICTED: ras-related protein RABE1a [Fragaria vesca subsp. vesca] |