| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
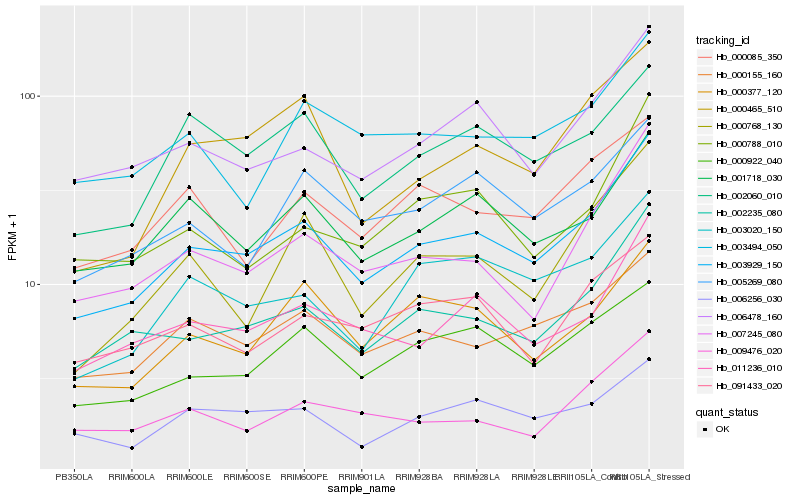
Hb_003929_150 |
0.0 |
- |
- |
PREDICTED: rRNA-processing protein fcf2-like [Jatropha curcas] |
| 2 |
Hb_002235_080 |
0.0752229213 |
- |
- |
PREDICTED: U3 small nucleolar ribonucleoprotein protein IMP3 [Vitis vinifera] |
| 3 |
Hb_011236_010 |
0.0894012892 |
- |
- |
hypothetical protein JCGZ_17006 [Jatropha curcas] |
| 4 |
Hb_006478_160 |
0.0920678691 |
- |
- |
20S proteasome beta subunit D1 [Hevea brasiliensis] |
| 5 |
Hb_000155_160 |
0.0955472343 |
- |
- |
hypothetical protein POPTR_0011s05880g [Populus trichocarpa] |
| 6 |
Hb_007245_080 |
0.0968020052 |
- |
- |
PREDICTED: uncharacterized protein LOC105644587 [Jatropha curcas] |
| 7 |
Hb_001718_030 |
0.0993738529 |
- |
- |
PREDICTED: calcineurin B-like protein 9 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000922_040 |
0.1002449822 |
- |
- |
PREDICTED: CASP-like protein 4A3 [Jatropha curcas] |
| 9 |
Hb_003020_150 |
0.1037846018 |
- |
- |
tropinone reductase, putative [Ricinus communis] |
| 10 |
Hb_006256_030 |
0.1046917057 |
- |
- |
PREDICTED: putative cyclic nucleotide-gated ion channel 18 [Jatropha curcas] |
| 11 |
Hb_005269_080 |
0.1068150301 |
- |
- |
protein with unknown function [Ricinus communis] |
| 12 |
Hb_003494_050 |
0.1078580207 |
- |
- |
hypothetical protein PRUPE_ppa013459mg [Prunus persica] |
| 13 |
Hb_000377_120 |
0.1082637815 |
- |
- |
PREDICTED: acyl carrier protein 3, mitochondrial isoform X2 [Jatropha curcas] |
| 14 |
Hb_000788_010 |
0.108895119 |
- |
- |
40S ribosomal protein S16A [Hevea brasiliensis] |
| 15 |
Hb_009476_020 |
0.1090664449 |
- |
- |
tryptophan biosynthesis protein, putative [Ricinus communis] |
| 16 |
Hb_002060_010 |
0.1092988226 |
- |
- |
PREDICTED: uncharacterized protein LOC105642435 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000465_510 |
0.1100573679 |
- |
- |
PREDICTED: GTP-binding protein YPTM2 [Populus euphratica] |
| 18 |
Hb_000768_130 |
0.1105290247 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
PREDICTED: protein TRI1 [Jatropha curcas] |
| 19 |
Hb_000085_350 |
0.1109862864 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM22-2-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_091433_020 |
0.111589242 |
- |
- |
ribosomal RNA methyltransferase, putative [Ricinus communis] |