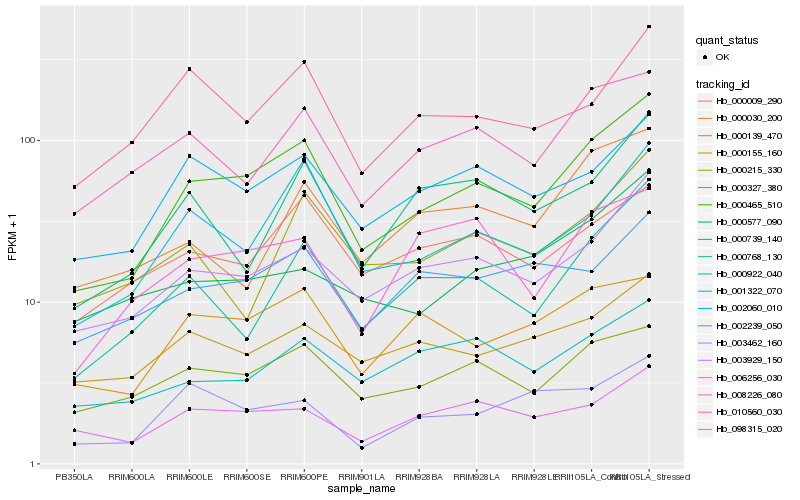
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000465_510 |
0.0 |
- |
- |
PREDICTED: GTP-binding protein YPTM2 [Populus euphratica] |
| 2 |
Hb_003929_150 |
0.1100573679 |
- |
- |
PREDICTED: rRNA-processing protein fcf2-like [Jatropha curcas] |
| 3 |
Hb_001322_070 |
0.1191940643 |
- |
- |
protein with unknown function [Ricinus communis] |
| 4 |
Hb_000768_130 |
0.1290989926 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
PREDICTED: protein TRI1 [Jatropha curcas] |
| 5 |
Hb_002060_010 |
0.1292879872 |
- |
- |
PREDICTED: uncharacterized protein LOC105642435 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000327_380 |
0.1330977953 |
- |
- |
hypothetical protein JCGZ_06621 [Jatropha curcas] |
| 7 |
Hb_010560_030 |
0.1364184073 |
- |
- |
PREDICTED: mitochondrial pyruvate carrier 4-like [Jatropha curcas] |
| 8 |
Hb_000009_290 |
0.141783556 |
- |
- |
PREDICTED: cold-regulated 413 plasma membrane protein 4-like [Jatropha curcas] |
| 9 |
Hb_000139_470 |
0.1427563494 |
- |
- |
Dual specificity protein phosphatase, putative [Ricinus communis] |
| 10 |
Hb_006256_030 |
0.1443094114 |
- |
- |
PREDICTED: putative cyclic nucleotide-gated ion channel 18 [Jatropha curcas] |
| 11 |
Hb_098315_020 |
0.1443199327 |
- |
- |
hypothetical protein M569_15712 [Genlisea aurea] |
| 12 |
Hb_000155_160 |
0.1452203525 |
- |
- |
hypothetical protein POPTR_0011s05880g [Populus trichocarpa] |
| 13 |
Hb_000739_140 |
0.1490746091 |
- |
- |
PREDICTED: proteasome subunit beta type-1 [Jatropha curcas] |
| 14 |
Hb_000922_040 |
0.1505821856 |
- |
- |
PREDICTED: CASP-like protein 4A3 [Jatropha curcas] |
| 15 |
Hb_002239_050 |
0.1522870789 |
- |
- |
PREDICTED: isovaleryl-CoA dehydrogenase, mitochondrial [Jatropha curcas] |
| 16 |
Hb_003462_160 |
0.1523036068 |
transcription factor |
TF Family: GNAT |
predicted protein [Arabidopsis lyrata subsp. lyrata] |
| 17 |
Hb_000215_330 |
0.153701511 |
- |
- |
PREDICTED: AP-4 complex subunit sigma [Jatropha curcas] |
| 18 |
Hb_000030_200 |
0.1542872223 |
- |
- |
PREDICTED: early nodulin-93-like [Populus euphratica] |
| 19 |
Hb_000577_090 |
0.1547821175 |
- |
- |
PREDICTED: 50S ribosomal protein L12-1, chloroplastic-like [Jatropha curcas] |
| 20 |
Hb_008226_080 |
0.1550528197 |
- |
- |
Protein LRP16, putative [Ricinus communis] |