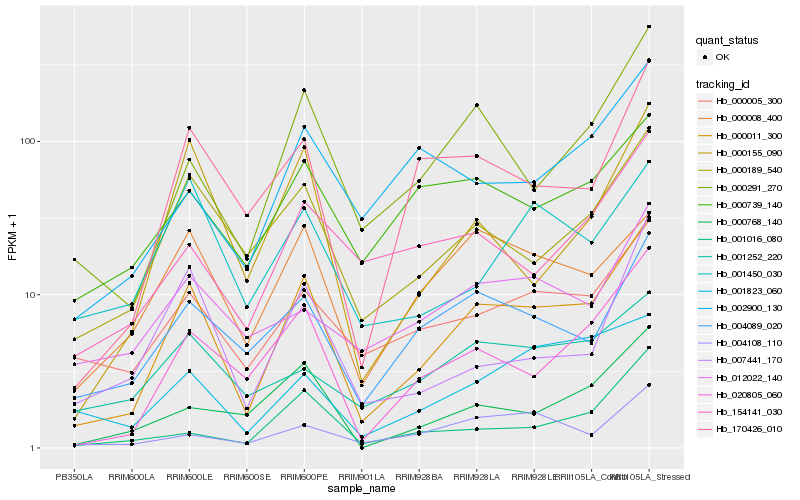
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000011_300 |
0.0 |
- |
- |
PREDICTED: proteasome subunit alpha type-5 isoform X1 [Gossypium raimondii] |
| 2 |
Hb_020805_060 |
0.1464786816 |
- |
- |
PREDICTED: protein S-acyltransferase 21 [Jatropha curcas] |
| 3 |
Hb_000189_540 |
0.1581327518 |
- |
- |
PREDICTED: tRNA(adenine(34)) deaminase, chloroplastic [Jatropha curcas] |
| 4 |
Hb_170426_010 |
0.1683040563 |
- |
- |
cytochrome b5 isoform Cb5-B [Vernicia fordii] |
| 5 |
Hb_000291_270 |
0.169373993 |
- |
- |
hypothetical protein JCGZ_12006 [Jatropha curcas] |
| 6 |
Hb_004089_020 |
0.1772842935 |
- |
- |
PREDICTED: 3-phosphoinositide-dependent protein kinase 2 [Jatropha curcas] |
| 7 |
Hb_000005_300 |
0.1799255458 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000155_090 |
0.1822194198 |
- |
- |
PREDICTED: glutaredoxin-C9 [Jatropha curcas] |
| 9 |
Hb_000768_140 |
0.186787726 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
tRNA (guanine-n(7)-)-methyltransferase, putative [Ricinus communis] |
| 10 |
Hb_000008_400 |
0.1976650653 |
- |
- |
PREDICTED: classical arabinogalactan protein 10-like [Nicotiana tomentosiformis] |
| 11 |
Hb_007441_170 |
0.1981212595 |
- |
- |
PREDICTED: rac-like GTP-binding protein RAC2 [Jatropha curcas] |
| 12 |
Hb_154141_030 |
0.199321239 |
- |
- |
PREDICTED: histone H2AX [Populus euphratica] |
| 13 |
Hb_000739_140 |
0.200427796 |
- |
- |
PREDICTED: proteasome subunit beta type-1 [Jatropha curcas] |
| 14 |
Hb_001016_080 |
0.2033132359 |
- |
- |
hypothetical protein EUGRSUZ_J02452 [Eucalyptus grandis] |
| 15 |
Hb_001823_060 |
0.205229165 |
- |
- |
fructose-bisphosphate aldolase, putative [Ricinus communis] |
| 16 |
Hb_001450_030 |
0.2074127603 |
- |
- |
PREDICTED: uncharacterized protein LOC105650827 [Jatropha curcas] |
| 17 |
Hb_012022_140 |
0.2076233984 |
- |
- |
ADP,ATP carrier protein, putative [Ricinus communis] |
| 18 |
Hb_001252_220 |
0.207929736 |
- |
- |
PREDICTED: uncharacterized protein LOC105640647 [Jatropha curcas] |
| 19 |
Hb_004108_110 |
0.2081851548 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_002900_130 |
0.212921584 |
- |
- |
60S ribosomal L35a-3 -like protein [Gossypium arboreum] |