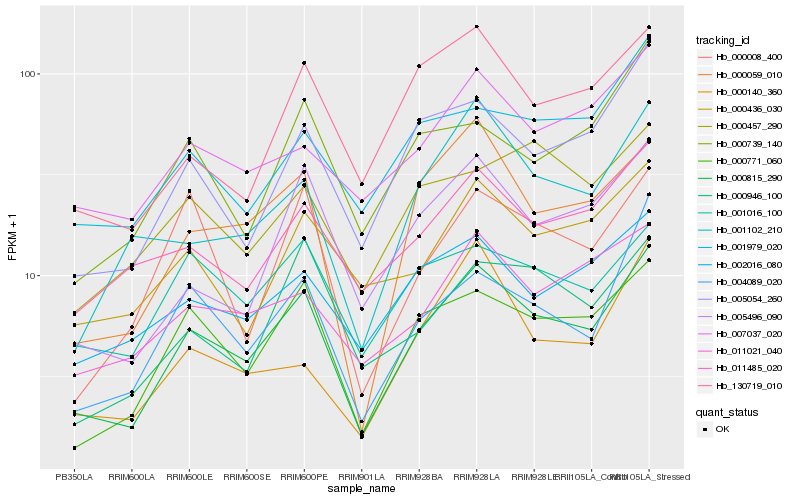
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000815_290 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000771_060 |
0.1011474038 |
- |
- |
PREDICTED: spermine synthase [Jatropha curcas] |
| 3 |
Hb_000059_010 |
0.1111624342 |
- |
- |
amino acid binding protein, putative [Ricinus communis] |
| 4 |
Hb_005054_260 |
0.1199591689 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 7 homolog A [Jatropha curcas] |
| 5 |
Hb_005496_090 |
0.1225457153 |
- |
- |
PREDICTED: adenine phosphoribosyltransferase 1-like [Jatropha curcas] |
| 6 |
Hb_004089_020 |
0.1249596888 |
- |
- |
PREDICTED: 3-phosphoinositide-dependent protein kinase 2 [Jatropha curcas] |
| 7 |
Hb_130719_010 |
0.1318804556 |
- |
- |
V-type proton ATPase subunit E [Hevea brasiliensis] |
| 8 |
Hb_001102_210 |
0.1333487979 |
- |
- |
PREDICTED: alpha,alpha-trehalose-phosphate synthase [UDP-forming] 1-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_011485_020 |
0.1347194857 |
- |
- |
protein with unknown function [Ricinus communis] |
| 10 |
Hb_002016_080 |
0.1361559854 |
- |
- |
PREDICTED: thiamine pyrophosphokinase 1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_011021_040 |
0.1375219118 |
transcription factor |
TF Family: Trihelix |
PREDICTED: phosphatidylinositol/phosphatidylcholine transfer protein SFH9 [Jatropha curcas] |
| 12 |
Hb_001016_100 |
0.1384170961 |
- |
- |
PREDICTED: V-type proton ATPase subunit E-like [Pyrus x bretschneideri] |
| 13 |
Hb_000946_100 |
0.1387013344 |
- |
- |
PREDICTED: SKI/DACH domain-containing protein 1-like [Jatropha curcas] |
| 14 |
Hb_000436_030 |
0.1396822387 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_007037_020 |
0.1407596596 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 78 [Jatropha curcas] |
| 16 |
Hb_000739_140 |
0.1438245859 |
- |
- |
PREDICTED: proteasome subunit beta type-1 [Jatropha curcas] |
| 17 |
Hb_000008_400 |
0.1444600053 |
- |
- |
PREDICTED: classical arabinogalactan protein 10-like [Nicotiana tomentosiformis] |
| 18 |
Hb_001979_020 |
0.1446954982 |
- |
- |
hypothetical protein OsJ_31823 [Oryza sativa Japonica Group] |
| 19 |
Hb_000457_290 |
0.1537875122 |
- |
- |
ATP-dependent Clp protease proteolytic subunit, putative [Ricinus communis] |
| 20 |
Hb_000140_360 |
0.1539122153 |
- |
- |
PREDICTED: uncharacterized protein LOC105642906 [Jatropha curcas] |