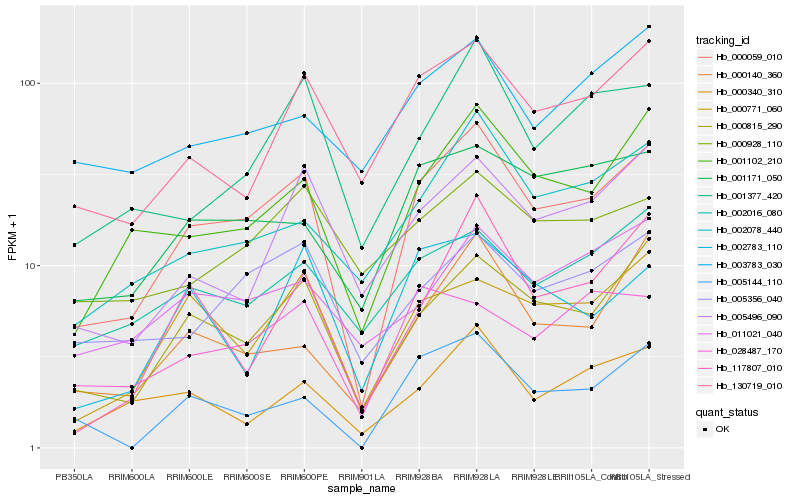
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000059_010 |
0.0 |
- |
- |
amino acid binding protein, putative [Ricinus communis] |
| 2 |
Hb_000815_290 |
0.1111624342 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X2 [Jatropha curcas] |
| 3 |
Hb_001102_210 |
0.1279780524 |
- |
- |
PREDICTED: alpha,alpha-trehalose-phosphate synthase [UDP-forming] 1-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_130719_010 |
0.1474316493 |
- |
- |
V-type proton ATPase subunit E [Hevea brasiliensis] |
| 5 |
Hb_002078_440 |
0.1512982368 |
transcription factor |
TF Family: Orphans |
PREDICTED: histidine kinase 3 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001377_420 |
0.1514685063 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000771_060 |
0.1519878466 |
- |
- |
PREDICTED: spermine synthase [Jatropha curcas] |
| 8 |
Hb_005356_040 |
0.1557390488 |
- |
- |
receptor serine-threonine protein kinase, putative [Ricinus communis] |
| 9 |
Hb_001171_050 |
0.1582349878 |
- |
- |
PREDICTED: uncharacterized protein DDB_G0284459 [Jatropha curcas] |
| 10 |
Hb_005496_090 |
0.1599814447 |
- |
- |
PREDICTED: adenine phosphoribosyltransferase 1-like [Jatropha curcas] |
| 11 |
Hb_002016_080 |
0.1644240759 |
- |
- |
PREDICTED: thiamine pyrophosphokinase 1 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000340_310 |
0.1644277277 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase RF298 [Jatropha curcas] |
| 13 |
Hb_005144_110 |
0.1648385489 |
- |
- |
PREDICTED: uncharacterized protein LOC104455487 [Eucalyptus grandis] |
| 14 |
Hb_000140_360 |
0.1650124985 |
- |
- |
PREDICTED: uncharacterized protein LOC105642906 [Jatropha curcas] |
| 15 |
Hb_117807_010 |
0.166365576 |
- |
- |
PREDICTED: uncharacterized protein LOC105635720 [Jatropha curcas] |
| 16 |
Hb_011021_040 |
0.1690997486 |
transcription factor |
TF Family: Trihelix |
PREDICTED: phosphatidylinositol/phosphatidylcholine transfer protein SFH9 [Jatropha curcas] |
| 17 |
Hb_002783_110 |
0.1710778694 |
- |
- |
Zeamatin precursor, putative [Ricinus communis] |
| 18 |
Hb_028487_170 |
0.1728571692 |
- |
- |
PREDICTED: uncharacterized protein LOC105634088 [Jatropha curcas] |
| 19 |
Hb_000928_110 |
0.1748573556 |
- |
- |
PREDICTED: uncharacterized protein LOC105632499 isoform X1 [Jatropha curcas] |
| 20 |
Hb_003783_030 |
0.1766546024 |
- |
- |
hypothetical protein POPTR_0018s13980g [Populus trichocarpa] |