| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
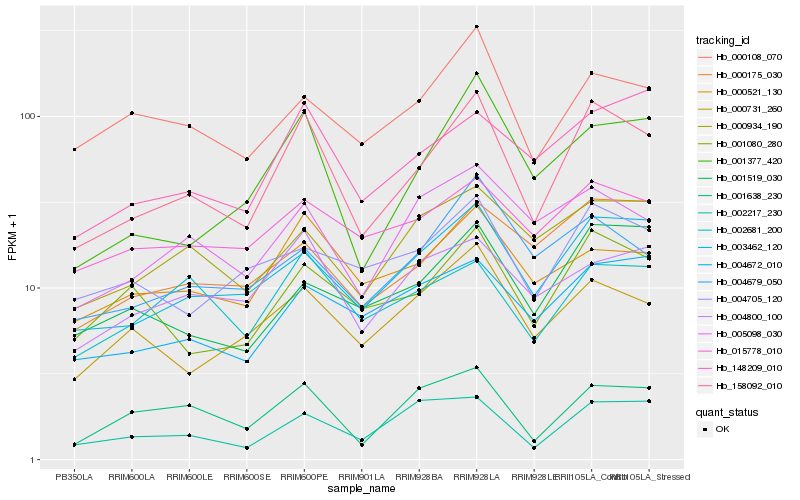
Hb_158092_010 |
0.0 |
- |
- |
CYP51 [Hevea brasiliensis] |
| 2 |
Hb_003462_120 |
0.1166340414 |
- |
- |
Acidic endochitinase [Theobroma cacao] |
| 3 |
Hb_001377_420 |
0.1285709692 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001080_280 |
0.1329558847 |
- |
- |
PREDICTED: uncharacterized protein LOC105649981 [Jatropha curcas] |
| 5 |
Hb_001638_230 |
0.1358292615 |
- |
- |
RNA-binding region-containing protein, putative [Ricinus communis] |
| 6 |
Hb_000731_260 |
0.1406180684 |
- |
- |
hypothetical protein RCOM_0905090 [Ricinus communis] |
| 7 |
Hb_148209_010 |
0.1430150042 |
- |
- |
RAS-related GTP-binding family protein [Populus trichocarpa] |
| 8 |
Hb_000521_130 |
0.1439257272 |
- |
- |
PREDICTED: citrate synthase, mitochondrial [Jatropha curcas] |
| 9 |
Hb_005098_030 |
0.1464522713 |
- |
- |
PREDICTED: argininosuccinate synthase, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000108_070 |
0.1478511586 |
- |
- |
alpha-tubulin 1 [Hevea brasiliensis] |
| 11 |
Hb_002217_230 |
0.1486030314 |
- |
- |
PREDICTED: uncharacterized protein LOC105643583 [Jatropha curcas] |
| 12 |
Hb_004679_050 |
0.1493116574 |
- |
- |
galactono-1,4-lactone dehydrogenase, putative [Ricinus communis] |
| 13 |
Hb_001519_030 |
0.149620019 |
- |
- |
PREDICTED: protein trichome birefringence-like 12 isoform X1 [Jatropha curcas] |
| 14 |
Hb_004800_100 |
0.1500053402 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000175_030 |
0.1510081211 |
- |
- |
Signal recognition particle, SRP54 subunit protein [Theobroma cacao] |
| 16 |
Hb_002681_200 |
0.1510313602 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RGLG2 [Jatropha curcas] |
| 17 |
Hb_004672_010 |
0.1513607397 |
- |
- |
ecotropic viral integration site, putative [Ricinus communis] |
| 18 |
Hb_000934_190 |
0.1522724168 |
- |
- |
SER/ARG-rich protein 34A [Theobroma cacao] |
| 19 |
Hb_004705_120 |
0.154221702 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 20 |
Hb_015778_010 |
0.1558574441 |
- |
- |
PREDICTED: AMSH-like ubiquitin thioesterase 1 [Jatropha curcas] |